Note
Go to the end to download the full example code.
The PET-MAD universal potential¶
- Authors:
Philip Loche @PicoCentauri, Michele Ceriotti @ceriottm, Arslan Mazitov @abmazitov
This example demonstrates how to use the PET-MAD model with ASE, i-PI and LAMMPS. PET-MAD is a “universal” machine-learning forcefield trained on a dataset that aims to incorporate a very high degree of structural diversity.
The point-edge transformer (PET) is an unconstrained architecture that achieves a high degree of symmetry compliance through data augmentation during training (see the PET paper for more details). The unconstrained nature of the model simplifies its implementation and structure, making it computationally efficient and very expressive.
The MAD dataset combines “stable” inorganic structures from the MC3D dataset, 2D structures from the MC2D dataset, and molecular crystals from the ShiftML dataset with “Maximum Atomic Diversity” configurations, generated by distorting the composition and structure of these stable templates. By doing so, PET-MAD achieves state-of-the-art accuracy despite the MAD dataset containing fewer than 100k structures. The reference DFT settings are highly converged, but limited to a PBEsol functional, so the accuracy against experimental data depends on how good this level of theory is for a given system. PET-MAD is introduced, and benchmarked for several challenging modeling tasks, in this preprint.
Start by importing the required libraries. To use PET-MAD, and obtain all the necessary dependencies, you can simply use pip to install the upet package:
pip install upet
import os
import subprocess
from copy import copy, deepcopy
# ASE and i-PI scripting utilities
import ase.units
import chemiscope
import matplotlib.pyplot as plt
# pet-mad ASE calculator
import numpy as np
import requests
import upet
from ase.optimize import LBFGS
from upet.calculator import UPETCalculator
from ipi.utils.mathtools import get_rotation_quadrature_lebedev
from ipi.utils.parsing import read_output, read_trajectory
from ipi.utils.scripting import (
InteractiveSimulation,
forcefield_xml,
motion_nvt_xml,
simulation_xml,
)
Inference on the MAD test set¶
We begin by using the ase-compatible calculator to evaluate energy and forces for
a test dataset that contains both hold-out structures from the MAD dataset, and a few
structures from popular datasets (MPtrj,
Alexandria,
OC2020,
SPICE,
MD22) re-computed
with consistent DFT settings.
Load the dataset¶
We fetch the dataset, and load only some of the structures, to speed up the example runtime on CPU. The model can also run (much faster) on GPUs if you have some at hand.
filename = "data/mad-test-mad-settings.xyz"
if not os.path.exists(filename):
url = (
"https://huggingface.co/lab-cosmo/pet-mad/resolve/"
"main/benchmarks/mad-test-mad-settings.xyz"
)
response = requests.get(url)
response.raise_for_status()
with open(filename, "wb") as f:
f.write(response.content)
test_structures = ase.io.read(filename, "::15")
# also extract reference energetics and metadata
test_energy = []
test_forces = []
test_natoms = []
test_origin = []
subsets = []
for s in test_structures:
test_energy.append(s.get_potential_energy())
test_natoms.append(len(s))
test_forces.append(s.get_forces())
test_origin.append(s.info["dataset"])
if s.info["dataset"] not in subsets:
subsets.append(s.info["dataset"])
test_natoms = np.array(test_natoms)
test_origin = np.array(test_origin)
test_energy = np.array(test_energy)
test_forces = np.array(test_forces, dtype=object)
Single point energy and forces¶
PET-MAD is compatible with the metatomic models interface which allows us to run it with ASE and many other MD engines. For more details see the metatomic documentation.
We now load the PET-MAD ASE calculator and calculate energy and forces.
We use the upet package, which provides a convenient interface
to download and use PET-MAD models. Here we use the small (s) model
at version 1.0.2, which was trained on PBEsol data matching the
reference level of theory of the MAD test set.
For general-purpose simulations, we recommend using version 1.5.0
(i.e. version="1.5.0") which is based on an improved architecture
and trained on r2SCAN meta-GGA references, unless PBEsol energetics is needed.
The extra-small (xs) model (model="pet-mad-xs", available from
version 1.5.0) is faster but less accurate than -s.
calculator = UPETCalculator(model="pet-mad-s", version="1.0.2", device="cpu")
The model can also be exported in a format that can be used with
external MD engines, such as i-PI and LAMMPS. This is done by saving
the model to a file using upet.save_upet.
model_path = "pet-mad-s-v1.0.2.pt"
upet.save_upet(model="pet-mad", size="s", version="1.0.2", output=model_path)
Here, we run the computation on the CPU. If you have a CUDA GPU you can also set
device="cuda" to speed up the computation.
mad_energy = []
mad_forces = []
mad_structures = []
for structure in test_structures:
tmp = deepcopy(structure)
tmp.calc = copy(calculator) # avoids overwriting results.
mad_energy.append(tmp.get_potential_energy())
mad_forces.append(tmp.get_forces())
mad_structures.append(tmp)
mad_energy = np.array(mad_energy)
mad_forces = np.array(mad_forces, dtype=object)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [2 3].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [0 1 2 3].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [36 37 38 39].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [2].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [0 1].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [4 5 6 7].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [26 27 28 29 30 31 32 33].
self.calculator.calculate(atoms, properties, system_changes)
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad/lib/python3.12/site-packages/upet/calculator.py:227: UserWarning: Some of the atomic energy uncertainties are larger than the threshold of 0.1 eV. The prediction is above the threshold for atoms [ 9 10 11 12 13 14 15].
self.calculator.calculate(atoms, properties, system_changes)
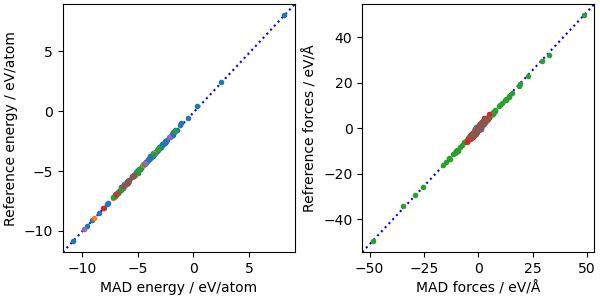
A parity plot with the model predictions
tab10 = plt.get_cmap("tab10")
fig, ax = plt.subplots(1, 2, figsize=(6, 3.5), constrained_layout=True)
ax[0].plot([0, 1], [0, 1], "b:", transform=ax[0].transAxes)
ax[1].plot([0, 1], [0, 1], "b:", transform=ax[1].transAxes)
for i, sub in enumerate(subsets):
sel = np.where(test_origin == sub)[0]
ax[0].plot(
mad_energy[sel] / test_natoms[sel],
test_energy[sel] / test_natoms[sel],
".",
c=tab10(i),
label=sub,
)
ax[1].plot(
np.concatenate(mad_forces[sel]).flatten(),
np.concatenate(test_forces[sel]).flatten(),
".",
c=tab10(i),
)
ax[0].set_xlabel("MAD energy / eV/atom")
ax[0].set_ylabel("Reference energy / eV/atom")
ax[1].set_xlabel("MAD forces / eV/Å")
ax[1].set_ylabel("Reference forces / eV/Å")
fig.legend(loc="outside upper center", ncol=3)
<matplotlib.legend.Legend object at 0x7f05b4e68ad0>
Explore the dataset using chemiscope
chemiscope.show(
test_structures,
mode="default",
properties={
"origin": test_origin,
"energy_ref": {
"target": "structure",
"values": test_energy / test_natoms,
"units": "eV/atom",
},
"energy_mad": {
"target": "structure",
"values": mad_energy / test_natoms,
"units": "eV/atom",
},
"energy_error": {
"target": "structure",
"values": np.abs((test_energy - mad_energy) / test_natoms),
"units": "eV/atom",
},
"force_error": {
"target": "structure",
"values": [
np.linalg.norm(f1 - f2) / n
for (f1, f2, n) in zip(mad_forces, test_forces, test_natoms)
],
"units": "eV/Å",
},
},
shapes={
"forces_ref": chemiscope.ase_vectors_to_arrows(
mad_structures, "forces", scale=5
),
"forces_mad": chemiscope.ase_vectors_to_arrows(
test_structures, "forces", scale=5
),
},
settings=chemiscope.quick_settings(
x="energy_mad",
y="energy_ref",
symbol="origin",
structure_settings={"unitCell": True, "shape": ["forces_ref"]},
),
)
How about equivariance‽¶
The PET architecture does not provide “intrinsically” invariant energy predictions, but learns symmetry from data augmentation. Should you worry? The authors of PET-MAD certainly do, and they have studied extensively whether the symmetry breaking can cause serious artefacts. You can check by yourself following the procedure below, that evaluates a structures over a grid of rotations, estimating the variability in energy (which is around 1meV/atom, much smaller than the test error).
rotations = get_rotation_quadrature_lebedev(3)
rot_test = test_structures[100]
rot_structures = []
rot_weights = []
rot_energies = []
rot_forces = []
rot_angles = []
for rot, w, angles in rotations:
tmp = rot_test.copy()
tmp.positions = tmp.positions @ rot.T
tmp.cell = tmp.cell @ rot.T
tmp.calc = copy(calculator)
rot_weights.append(w)
rot_energies.append(tmp.get_potential_energy() / len(tmp))
rot_forces.append(tmp.get_forces())
rot_structures.append(tmp)
rot_angles.append(angles)
rot_energies = np.array(rot_energies)
rot_weights = np.array(rot_weights)
rot_angles = np.array(rot_angles)
erot_rms = 1e3 * np.sqrt(
np.sum(rot_energies**2 * rot_weights) / np.sum(rot_weights)
- (np.sum(rot_energies * rot_weights) / np.sum(rot_weights)) ** 2
)
erot_max = 1e3 * np.abs(rot_energies.max() - rot_energies.min())
print(f"""
Symmetry breaking, energy:
RMS: {erot_rms:.3f} meV/at.
Max: {erot_max:.3f} meV/at.
""")
Symmetry breaking, energy:
RMS: 0.283 meV/at.
Max: 1.004 meV/at.
You can also inspect the rotational behavior visually
chemiscope.show(
rot_structures,
mode="default",
properties={
"delta_energy": {
"target": "structure",
"values": 1e3 * (rot_energies - rot_energies.mean()),
"units": "meV/atom",
},
"euler_angles": rot_angles,
},
shapes={
"forces": chemiscope.ase_vectors_to_arrows(rot_structures, "forces", scale=1),
},
settings=chemiscope.quick_settings(
x="euler_angles[1]",
y="euler_angles[2]",
z="euler_angles[3]",
map_color="delta_energy",
structure_settings={"unitCell": True, "shape": ["forces"]},
),
)
Note also that i-PI provides functionalities to do this automatically to obtain MD trajectories with a even higher degree of symmetry-compliance.
Simulating a complex surface¶
PET-MAD is designed to be robust and stable when executing sophisticated modeling workflows. As an example, we consider a slab of an Al-6xxx alloy (aluminum with a few percent Mg and Si) with some oxygen molecules adsorbed at the (111) surface.
Warning
The overall Si+Mg concentration in an Al6xxx alloy is far lower than what depicted here. This is just a demonstrative example and should not be taken as the starting point of a serious study of this system.
al_surface = ase.io.read("data/al6xxx-o2.xyz")
Geometry optimization with ASE¶
As a first example, we use the ase geometry LBFGS optimizer to relax the initial
positions. This leads to the rapid decomposition of the oxygen molecules and the
formation of an oxide layer.
atoms = al_surface.copy()
atoms.calc = calculator
opt = LBFGS(atoms)
traj_atoms = []
traj_energy = []
opt.attach(lambda: traj_atoms.append(atoms.copy()))
opt.attach(lambda: traj_energy.append(atoms.get_potential_energy()))
# stop the optimization early to speed up the example
opt.run(fmax=0.001, steps=20)
Step Time Energy fmax
LBFGS: 0 06:52:12 -756.134521 3.571309
LBFGS: 1 06:52:12 -758.695557 2.791725
LBFGS: 2 06:52:13 -759.814514 9.726063
LBFGS: 3 06:52:13 -762.155579 1.981836
LBFGS: 4 06:52:14 -762.748413 1.932890
LBFGS: 5 06:52:14 -763.676208 2.221239
LBFGS: 6 06:52:14 -764.275085 1.837465
LBFGS: 7 06:52:15 -765.156311 3.671942
LBFGS: 8 06:52:15 -765.424683 1.181422
LBFGS: 9 06:52:15 -765.619202 1.046657
LBFGS: 10 06:52:16 -766.063965 1.444244
LBFGS: 11 06:52:16 -766.304871 1.109468
LBFGS: 12 06:52:16 -766.739380 1.491528
LBFGS: 13 06:52:17 -766.981079 1.742016
LBFGS: 14 06:52:17 -767.375122 1.701642
LBFGS: 15 06:52:17 -767.819702 1.461242
LBFGS: 16 06:52:18 -768.149597 1.670298
LBFGS: 17 06:52:18 -768.566895 1.355886
LBFGS: 18 06:52:19 -769.125488 1.679120
LBFGS: 19 06:52:19 -769.680054 1.669325
LBFGS: 20 06:52:19 -770.123779 1.823701
np.False_
Even if the optimization is cut short and far from converged, the decomposition of the oxygen molecules is apparent, and leads to a large energetic stabilization
chemiscope.show(
structures=traj_atoms,
properties={
"index": np.arange(0, len(traj_atoms)),
"energy": traj_energy,
},
mode="default",
settings=chemiscope.quick_settings(
trajectory=True,
structure_settings={
"unitCell": True,
},
),
)
Molecular dynamics with atoms exchange with i-PI¶
The geometry optimization shows the high reactivity of this surface, but does not properly account for finite temperature and does not sample the diffusion of solute atoms in the alloy (which is mediated by vacancies).
We use i-PI to perform a molecular dynamics trajectory at 800K, combined with Monte Carlo steps that swap the nature of atoms, allowing the simulation to reach equilibrium in the solute-atoms distributions without having to introduce vacancies or wait for the very long time scale needed for diffusion.
The behavior of i-PI is controlled by an XML input file.
The utils.scripting module contains several helper
functions to generate the basic components.
Here we use a <motion mode="multi"> block to combine
a MD run with a <motion mode="atomswap"> block that
attempts swapping atoms, with a Monte Carlo acceptance.
motion_xml = f"""
<motion mode="multi">
{motion_nvt_xml(timestep=5.0 * ase.units.fs)}
<motion mode="atomswap">
<atomswap>
<nxc> 0.1 </nxc>
<names> [ Al, Si, Mg, O] </names>
</atomswap>
</motion>
</motion>
"""
input_xml = simulation_xml(
structures=[al_surface],
forcefield=forcefield_xml(
name="pet-mad",
mode="direct",
pes="metatomic",
parameters={"model": "pet-mad-s-v1.0.2.pt", "template": "data/al6xxx-o2.xyz"},
),
motion=motion_xml,
temperature=800,
verbosity="high",
prefix="nvt_atomxc",
)
print(input_xml)
<simulation verbosity='high' safe_stride='20'>
<ffdirect name='pet-mad'>
<pes>metatomic</pes>
<parameters>{model: pet-mad-s-v1.0.2.pt, template: data/al6xxx-o2.xyz}</parameters>
</ffdirect>
<output prefix='nvt_atomxc'>
<properties stride='2' filename='out'>[ step, time{picosecond}, conserved{electronvolt}, temperature{kelvin}, potential{electronvolt} ]</properties>
<trajectory filename='pos' stride='20' cell_units='angstrom'>positions{angstrom}</trajectory>
<checkpoint stride='200'>
</checkpoint>
</output>
<system>
<beads natoms='191' nbeads='1'>
<q shape='(1, 573)'>
[ 0.00000000e+00, 0.00000000e+00, 2.77346357e+01, 5.41176453e+00, 0.00000000e+00,
2.77346357e+01, 1.08235291e+01, 0.00000000e+00, 2.77346357e+01, 1.62352936e+01,
0.00000000e+00, 2.77346357e+01, 2.16470582e+01, 0.00000000e+00, 2.77346357e+01,
2.70588227e+00, 4.68672556e+00, 2.77346357e+01, 8.11764682e+00, 4.68672556e+00,
2.77346357e+01, 1.35294114e+01, 4.68672556e+00, 2.77346357e+01, 1.89411759e+01,
4.68672556e+00, 2.77346357e+01, 2.43529404e+01, 4.68672556e+00, 2.77346357e+01,
5.41176453e+00, 9.37345114e+00, 2.77346357e+01, 1.08235291e+01, 9.37345114e+00,
2.77346357e+01, 1.62352936e+01, 9.37345114e+00, 2.77346357e+01, 2.16470582e+01,
9.37345114e+00, 2.77346357e+01, 2.70588227e+01, 9.37345114e+00, 2.77346357e+01,
8.11764682e+00, 1.40601767e+01, 2.77346357e+01, 1.35294114e+01, 1.40601767e+01,
2.77346357e+01, 1.89411759e+01, 1.40601767e+01, 2.77346357e+01, 2.43529404e+01,
1.40601767e+01, 2.77346357e+01, 2.97647050e+01, 1.40601767e+01, 2.77346357e+01,
1.08235291e+01, 1.87469023e+01, 2.77346357e+01, 1.62352936e+01, 1.87469023e+01,
2.77346357e+01, 2.16470582e+01, 1.87469023e+01, 2.77346357e+01, 2.70588227e+01,
1.87469023e+01, 2.77346357e+01, 3.24705872e+01, 1.87469023e+01, 2.77346357e+01,
2.70588227e+00, 1.56224186e+00, 3.21533230e+01, 8.11764682e+00, 1.56224186e+00,
3.21533230e+01, 1.35294114e+01, 1.56224186e+00, 3.21533230e+01, 1.89411759e+01,
1.56224186e+00, 3.21533230e+01, 2.43529404e+01, 1.56224186e+00, 3.21533230e+01,
5.41176453e+00, 6.24896742e+00, 3.21533230e+01, 1.08235291e+01, 6.24896742e+00,
3.21533230e+01, 1.62352936e+01, 6.24896742e+00, 3.21533230e+01, 2.16470582e+01,
6.24896742e+00, 3.21533230e+01, 2.70588227e+01, 6.24896742e+00, 3.21533230e+01,
8.11764682e+00, 1.09356930e+01, 3.21533230e+01, 1.35294114e+01, 1.09356930e+01,
3.21533230e+01, 1.89411759e+01, 1.09356930e+01, 3.21533230e+01, 2.43529404e+01,
1.09356930e+01, 3.21533230e+01, 2.97647050e+01, 1.09356930e+01, 3.21533230e+01,
1.08235291e+01, 1.56224186e+01, 3.21533230e+01, 1.62352936e+01, 1.56224186e+01,
3.21533230e+01, 2.16470582e+01, 1.56224186e+01, 3.21533230e+01, 2.70588227e+01,
1.56224186e+01, 3.21533230e+01, 3.24705872e+01, 1.56224186e+01, 3.21533230e+01,
1.35294114e+01, 2.03091441e+01, 3.21533230e+01, 1.89411759e+01, 2.03091441e+01,
3.21533230e+01, 2.43529404e+01, 2.03091441e+01, 3.21533230e+01, 2.97647050e+01,
2.03091441e+01, 3.21533230e+01, 3.51764695e+01, 2.03091441e+01, 3.21533230e+01,
-0.00000000e+00, 3.12448372e+00, 3.65720102e+01, 5.41176453e+00, 3.12448372e+00,
3.65720102e+01, 1.08235291e+01, 3.12448372e+00, 3.65720102e+01, 1.62352936e+01,
3.12448372e+00, 3.65720102e+01, 2.16470582e+01, 3.12448372e+00, 3.65720102e+01,
2.70588227e+00, 7.81120928e+00, 3.65720102e+01, 8.11764682e+00, 7.81120928e+00,
3.65720102e+01, 1.35294114e+01, 7.81120928e+00, 3.65720102e+01, 1.89411759e+01,
7.81120928e+00, 3.65720102e+01, 2.43529404e+01, 7.81120928e+00, 3.65720102e+01,
5.41176453e+00, 1.24979349e+01, 3.65720102e+01, 1.08235291e+01, 1.24979349e+01,
3.65720102e+01, 1.62352936e+01, 1.24979349e+01, 3.65720102e+01, 2.16470582e+01,
1.24979349e+01, 3.65720102e+01, 2.70588227e+01, 1.24979349e+01, 3.65720102e+01,
8.11764682e+00, 1.71846604e+01, 3.65720102e+01, 1.35294114e+01, 1.71846604e+01,
3.65720102e+01, 1.89411759e+01, 1.71846604e+01, 3.65720102e+01, 2.43529404e+01,
1.71846604e+01, 3.65720102e+01, 2.97647050e+01, 1.71846604e+01, 3.65720102e+01,
1.08235291e+01, 2.18713860e+01, 3.65720102e+01, 1.62352936e+01, 2.18713860e+01,
3.65720102e+01, 2.16470582e+01, 2.18713860e+01, 3.65720102e+01, 2.70588227e+01,
2.18713860e+01, 3.65720102e+01, 3.24705872e+01, 2.18713860e+01, 3.65720102e+01,
0.00000000e+00, 0.00000000e+00, 4.09906975e+01, 5.41176453e+00, 0.00000000e+00,
4.09906975e+01, 1.08235291e+01, 0.00000000e+00, 4.09906975e+01, 1.62352936e+01,
0.00000000e+00, 4.09906975e+01, 2.16470582e+01, 0.00000000e+00, 4.09906975e+01,
2.70588227e+00, 4.68672556e+00, 4.09906975e+01, 8.11764682e+00, 4.68672556e+00,
4.09906975e+01, 1.35294114e+01, 4.68672556e+00, 4.09906975e+01, 1.89411759e+01,
4.68672556e+00, 4.09906975e+01, 2.43529404e+01, 4.68672556e+00, 4.09906975e+01,
5.41176453e+00, 9.37345114e+00, 4.09906975e+01, 1.08235291e+01, 9.37345114e+00,
4.09906975e+01, 1.62352936e+01, 9.37345114e+00, 4.09906975e+01, 2.16470582e+01,
9.37345114e+00, 4.09906975e+01, 2.70588227e+01, 9.37345114e+00, 4.09906975e+01,
8.11764682e+00, 1.40601767e+01, 4.09906975e+01, 1.35294114e+01, 1.40601767e+01,
4.09906975e+01, 1.89411759e+01, 1.40601767e+01, 4.09906975e+01, 2.43529404e+01,
1.40601767e+01, 4.09906975e+01, 2.97647050e+01, 1.40601767e+01, 4.09906975e+01,
1.08235291e+01, 1.87469023e+01, 4.09906975e+01, 1.62352936e+01, 1.87469023e+01,
4.09906975e+01, 2.16470582e+01, 1.87469023e+01, 4.09906975e+01, 2.70588227e+01,
1.87469023e+01, 4.09906975e+01, 3.24705872e+01, 1.87469023e+01, 4.09906975e+01,
2.70588227e+00, 1.56224186e+00, 4.54093847e+01, 8.11764682e+00, 1.56224186e+00,
4.54093847e+01, 1.35294114e+01, 1.56224186e+00, 4.54093847e+01, 1.89411759e+01,
1.56224186e+00, 4.54093847e+01, 2.43529404e+01, 1.56224186e+00, 4.54093847e+01,
5.41176453e+00, 6.24896742e+00, 4.54093847e+01, 1.08235291e+01, 6.24896742e+00,
4.54093847e+01, 1.62352936e+01, 6.24896742e+00, 4.54093847e+01, 2.16470582e+01,
6.24896742e+00, 4.54093847e+01, 2.70588227e+01, 6.24896742e+00, 4.54093847e+01,
8.11764682e+00, 1.09356930e+01, 4.54093847e+01, 1.35294114e+01, 1.09356930e+01,
4.54093847e+01, 1.89411759e+01, 1.09356930e+01, 4.54093847e+01, 2.43529404e+01,
1.09356930e+01, 4.54093847e+01, 2.97647050e+01, 1.09356930e+01, 4.54093847e+01,
1.08235291e+01, 1.56224186e+01, 4.54093847e+01, 1.62352936e+01, 1.56224186e+01,
4.54093847e+01, 2.16470582e+01, 1.56224186e+01, 4.54093847e+01, 2.70588227e+01,
1.56224186e+01, 4.54093847e+01, 3.24705872e+01, 1.56224186e+01, 4.54093847e+01,
1.35294114e+01, 2.03091441e+01, 4.54093847e+01, 1.89411759e+01, 2.03091441e+01,
4.54093847e+01, 2.43529404e+01, 2.03091441e+01, 4.54093847e+01, 2.97647050e+01,
2.03091441e+01, 4.54093847e+01, 3.51764695e+01, 2.03091441e+01, 4.54093847e+01,
-0.00000000e+00, 3.12448372e+00, 4.98280720e+01, 5.41176453e+00, 3.12448372e+00,
4.98280720e+01, 1.08235291e+01, 3.12448372e+00, 4.98280720e+01, 1.62352936e+01,
3.12448372e+00, 4.98280720e+01, 2.16470582e+01, 3.12448372e+00, 4.98280720e+01,
2.70588227e+00, 7.81120928e+00, 4.98280720e+01, 8.11764682e+00, 7.81120928e+00,
4.98280720e+01, 1.35294114e+01, 7.81120928e+00, 4.98280720e+01, 1.89411759e+01,
7.81120928e+00, 4.98280720e+01, 2.43529404e+01, 7.81120928e+00, 4.98280720e+01,
5.41176453e+00, 1.24979349e+01, 4.98280720e+01, 1.08235291e+01, 1.24979349e+01,
4.98280720e+01, 1.62352936e+01, 1.24979349e+01, 4.98280720e+01, 2.16470582e+01,
1.24979349e+01, 4.98280720e+01, 2.70588227e+01, 1.24979349e+01, 4.98280720e+01,
8.11764682e+00, 1.71846604e+01, 4.98280720e+01, 1.35294114e+01, 1.71846604e+01,
4.98280720e+01, 1.89411759e+01, 1.71846604e+01, 4.98280720e+01, 2.43529404e+01,
1.71846604e+01, 4.98280720e+01, 2.97647050e+01, 1.71846604e+01, 4.98280720e+01,
1.08235291e+01, 2.18713860e+01, 4.98280720e+01, 1.62352936e+01, 2.18713860e+01,
4.98280720e+01, 2.16470582e+01, 2.18713860e+01, 4.98280720e+01, 2.70588227e+01,
2.18713860e+01, 4.98280720e+01, 3.24705872e+01, 2.18713860e+01, 4.98280720e+01,
0.00000000e+00, 0.00000000e+00, 5.42467592e+01, 5.41176453e+00, 0.00000000e+00,
5.42467592e+01, 1.08235291e+01, 0.00000000e+00, 5.42467592e+01, 1.62352936e+01,
0.00000000e+00, 5.42467592e+01, 2.16470582e+01, 0.00000000e+00, 5.42467592e+01,
2.70588227e+00, 4.68672556e+00, 5.42467592e+01, 8.11764682e+00, 4.68672556e+00,
5.42467592e+01, 1.35294114e+01, 4.68672556e+00, 5.42467592e+01, 1.89411759e+01,
4.68672556e+00, 5.42467592e+01, 2.43529404e+01, 4.68672556e+00, 5.42467592e+01,
5.41176453e+00, 9.37345114e+00, 5.42467592e+01, 1.08235291e+01, 9.37345114e+00,
5.42467592e+01, 1.62352936e+01, 9.37345114e+00, 5.42467592e+01, 2.16470582e+01,
9.37345114e+00, 5.42467592e+01, 2.70588227e+01, 9.37345114e+00, 5.42467592e+01,
8.11764682e+00, 1.40601767e+01, 5.42467592e+01, 1.35294114e+01, 1.40601767e+01,
5.42467592e+01, 1.89411759e+01, 1.40601767e+01, 5.42467592e+01, 2.43529404e+01,
1.40601767e+01, 5.42467592e+01, 2.97647050e+01, 1.40601767e+01, 5.42467592e+01,
1.08235291e+01, 1.87469023e+01, 5.42467592e+01, 1.62352936e+01, 1.87469023e+01,
5.42467592e+01, 2.16470582e+01, 1.87469023e+01, 5.42467592e+01, 2.70588227e+01,
1.87469023e+01, 5.42467592e+01, 3.24705872e+01, 1.87469023e+01, 5.42467592e+01,
0.00000000e+00, 0.00000000e+00, 5.80262115e+01, -3.02356180e+00, 0.00000000e+00,
5.80262115e+01, 9.44863063e+00, 0.00000000e+00, 5.80262115e+01, 6.42506883e+00,
0.00000000e+00, 5.80262115e+01, 1.88972613e+01, 0.00000000e+00, 5.80262115e+01,
1.58736995e+01, 0.00000000e+00, 5.80262115e+01, 9.44863063e+00, 9.44863063e+00,
5.80262115e+01, 6.42506883e+00, 9.44863063e+00, 5.80262115e+01, 1.88972613e+01,
9.44863063e+00, 5.80262115e+01, 1.58736995e+01, 9.44863063e+00, 5.80262115e+01,
2.83458919e+01, 9.44863063e+00, 5.80262115e+01, 2.53223301e+01, 9.44863063e+00,
5.80262115e+01, 1.88972613e+01, 1.88972613e+01, 5.80262115e+01, 1.58736995e+01,
1.88972613e+01, 5.80262115e+01, 2.83458919e+01, 1.88972613e+01, 5.80262115e+01,
2.53223301e+01, 1.88972613e+01, 5.80262115e+01 ]
</q>
<p shape='(1, 573)'>
[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00 ]
</p>
<m shape='(191)'>
[ 4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 5.11967350e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04 ]
</m>
<names shape='(191)'>
[ Mg, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Si, Al, Mg, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Si,
Al, Al, Al, Al, Al,
Al, Al, Al, Mg, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Si, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Si, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Mg, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Si, Al, Al, Al,
Al, Al, Al, Al, Si,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Mg, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Al, Al, Al, Si,
O, O, O, O, O,
O, O, O, O, O,
O, O, O, O, O,
O ]
</names>
</beads>
<cell shape='(3, 3)'>
[ 2.70588227e+01, 1.35294114e+01, 0.00000000e+00, 0.00000000e+00, 2.34336279e+01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 7.31440205e+01 ]
</cell>
<initialize nbeads='1'><velocities mode='thermal' units='ase'> 800 </velocities></initialize><ensemble><temperature units='ase'> 800 </temperature></ensemble>
<forces>
<force forcefield='pet-mad'> </force>
</forces>
<motion mode="multi">
<motion mode="dynamics">
<dynamics mode="nvt">
<timestep units="ase"> 0.4911347394232032 </timestep>
<thermostat mode='svr'>
<tau units='ase'> 4.911347394232032 </tau>
</thermostat>
</dynamics>
</motion>
<motion mode="atomswap">
<atomswap>
<nxc> 0.1 </nxc>
<names> [ Al, Si, Mg, O] </names>
</atomswap>
</motion>
</motion>
</system>
</simulation>
The simulation can be run from a Python script or the command line. By changing the
forcefield interface from direct to the use of a socket, it is also possible to
execute separately i-PI and the metatomic driver.
sim = InteractiveSimulation(input_xml)
sim.run(80)
@system: Initializing system object
@simulation: Initializing simulation object
@ RANDOM SEED: The seed used in this calculation was 1772607140096
@initializer: Initializer (stage 1) parsing velocities object.
!W! Overwriting previous atomic momenta
@initializer: Resampling velocities at temperature 800.0 ase
--- begin input file content ---
<simulation verbosity='high' safe_stride='20'>
<ffdirect name='pet-mad'>
<pes>metatomic</pes>
<parameters>{model: pet-mad-s-v1.0.2.pt, template: data/al6xxx-o2.xyz}</parameters>
</ffdirect>
<output prefix='nvt_atomxc'>
<properties stride='2' filename='out'>[ step, time{picosecond}, conserved{electronvolt}, temperature{kelvin}, potential{electronvolt} ]</properties>
<trajectory filename='pos' stride='20' cell_units='angstrom'>positions{angstrom}</trajectory>
<checkpoint stride='200'>
</checkpoint>
</output>
<system>
<beads natoms='191' nbeads='1'>
<q shape='(1, 573)'>[ 0.00000000e+00, 0.00000000e+00, 2.77346357e+01, 5.41176453e+00, 0.00000000e+00,
2.77346357e+01, 1.08235291e+01, 0.00000000e+00, 2.77346357e+01, 1.62352936e+01,
0.00000000e+00, 2.77346357e+01, 2.16470582e+01, 0.00000000e+00, 2.77346357e+01,
2.70588227e+00, 4.68672556e+00, 2.77346357e+01, 8.11764682e+00, 4.68672556e+00,
2.77346357e+01, 1.35294114e+01, 4.68672556e+00, 2.77346357e+01, 1.89411759e+01,
4.68672556e+00, 2.77346357e+01, 2.43529404e+01, 4.68672556e+00, 2.77346357e+01,
5.41176453e+00, 9.37345114e+00, 2.77346357e+01, 1.08235291e+01, 9.37345114e+00,
2.77346357e+01, 1.62352936e+01, 9.37345114e+00, 2.77346357e+01, 2.16470582e+01,
9.37345114e+00, 2.77346357e+01, 2.70588227e+01, 9.37345114e+00, 2.77346357e+01,
8.11764682e+00, 1.40601767e+01, 2.77346357e+01, 1.35294114e+01, 1.40601767e+01,
2.77346357e+01, 1.89411759e+01, 1.40601767e+01, 2.77346357e+01, 2.43529404e+01,
1.40601767e+01, 2.77346357e+01, 2.97647050e+01, 1.40601767e+01, 2.77346357e+01,
1.08235291e+01, 1.87469023e+01, 2.77346357e+01, 1.62352936e+01, 1.87469023e+01,
2.77346357e+01, 2.16470582e+01, 1.87469023e+01, 2.77346357e+01, 2.70588227e+01,
1.87469023e+01, 2.77346357e+01, 3.24705872e+01, 1.87469023e+01, 2.77346357e+01,
2.70588227e+00, 1.56224186e+00, 3.21533230e+01, 8.11764682e+00, 1.56224186e+00,
3.21533230e+01, 1.35294114e+01, 1.56224186e+00, 3.21533230e+01, 1.89411759e+01,
1.56224186e+00, 3.21533230e+01, 2.43529404e+01, 1.56224186e+00, 3.21533230e+01,
5.41176453e+00, 6.24896742e+00, 3.21533230e+01, 1.08235291e+01, 6.24896742e+00,
3.21533230e+01, 1.62352936e+01, 6.24896742e+00, 3.21533230e+01, 2.16470582e+01,
6.24896742e+00, 3.21533230e+01, 2.70588227e+01, 6.24896742e+00, 3.21533230e+01,
8.11764682e+00, 1.09356930e+01, 3.21533230e+01, 1.35294114e+01, 1.09356930e+01,
3.21533230e+01, 1.89411759e+01, 1.09356930e+01, 3.21533230e+01, 2.43529404e+01,
1.09356930e+01, 3.21533230e+01, 2.97647050e+01, 1.09356930e+01, 3.21533230e+01,
1.08235291e+01, 1.56224186e+01, 3.21533230e+01, 1.62352936e+01, 1.56224186e+01,
3.21533230e+01, 2.16470582e+01, 1.56224186e+01, 3.21533230e+01, 2.70588227e+01,
1.56224186e+01, 3.21533230e+01, 3.24705872e+01, 1.56224186e+01, 3.21533230e+01,
1.35294114e+01, 2.03091441e+01, 3.21533230e+01, 1.89411759e+01, 2.03091441e+01,
3.21533230e+01, 2.43529404e+01, 2.03091441e+01, 3.21533230e+01, 2.97647050e+01,
2.03091441e+01, 3.21533230e+01, 3.51764695e+01, 2.03091441e+01, 3.21533230e+01,
-0.00000000e+00, 3.12448372e+00, 3.65720102e+01, 5.41176453e+00, 3.12448372e+00,
3.65720102e+01, 1.08235291e+01, 3.12448372e+00, 3.65720102e+01, 1.62352936e+01,
3.12448372e+00, 3.65720102e+01, 2.16470582e+01, 3.12448372e+00, 3.65720102e+01,
2.70588227e+00, 7.81120928e+00, 3.65720102e+01, 8.11764682e+00, 7.81120928e+00,
3.65720102e+01, 1.35294114e+01, 7.81120928e+00, 3.65720102e+01, 1.89411759e+01,
7.81120928e+00, 3.65720102e+01, 2.43529404e+01, 7.81120928e+00, 3.65720102e+01,
5.41176453e+00, 1.24979349e+01, 3.65720102e+01, 1.08235291e+01, 1.24979349e+01,
3.65720102e+01, 1.62352936e+01, 1.24979349e+01, 3.65720102e+01, 2.16470582e+01,
1.24979349e+01, 3.65720102e+01, 2.70588227e+01, 1.24979349e+01, 3.65720102e+01,
8.11764682e+00, 1.71846604e+01, 3.65720102e+01, 1.35294114e+01, 1.71846604e+01,
3.65720102e+01, 1.89411759e+01, 1.71846604e+01, 3.65720102e+01, 2.43529404e+01,
1.71846604e+01, 3.65720102e+01, 2.97647050e+01, 1.71846604e+01, 3.65720102e+01,
1.08235291e+01, 2.18713860e+01, 3.65720102e+01, 1.62352936e+01, 2.18713860e+01,
3.65720102e+01, 2.16470582e+01, 2.18713860e+01, 3.65720102e+01, 2.70588227e+01,
2.18713860e+01, 3.65720102e+01, 3.24705872e+01, 2.18713860e+01, 3.65720102e+01,
0.00000000e+00, 0.00000000e+00, 4.09906975e+01, 5.41176453e+00, 0.00000000e+00,
4.09906975e+01, 1.08235291e+01, 0.00000000e+00, 4.09906975e+01, 1.62352936e+01,
0.00000000e+00, 4.09906975e+01, 2.16470582e+01, 0.00000000e+00, 4.09906975e+01,
2.70588227e+00, 4.68672556e+00, 4.09906975e+01, 8.11764682e+00, 4.68672556e+00,
4.09906975e+01, 1.35294114e+01, 4.68672556e+00, 4.09906975e+01, 1.89411759e+01,
4.68672556e+00, 4.09906975e+01, 2.43529404e+01, 4.68672556e+00, 4.09906975e+01,
5.41176453e+00, 9.37345114e+00, 4.09906975e+01, 1.08235291e+01, 9.37345114e+00,
4.09906975e+01, 1.62352936e+01, 9.37345114e+00, 4.09906975e+01, 2.16470582e+01,
9.37345114e+00, 4.09906975e+01, 2.70588227e+01, 9.37345114e+00, 4.09906975e+01,
8.11764682e+00, 1.40601767e+01, 4.09906975e+01, 1.35294114e+01, 1.40601767e+01,
4.09906975e+01, 1.89411759e+01, 1.40601767e+01, 4.09906975e+01, 2.43529404e+01,
1.40601767e+01, 4.09906975e+01, 2.97647050e+01, 1.40601767e+01, 4.09906975e+01,
1.08235291e+01, 1.87469023e+01, 4.09906975e+01, 1.62352936e+01, 1.87469023e+01,
4.09906975e+01, 2.16470582e+01, 1.87469023e+01, 4.09906975e+01, 2.70588227e+01,
1.87469023e+01, 4.09906975e+01, 3.24705872e+01, 1.87469023e+01, 4.09906975e+01,
2.70588227e+00, 1.56224186e+00, 4.54093847e+01, 8.11764682e+00, 1.56224186e+00,
4.54093847e+01, 1.35294114e+01, 1.56224186e+00, 4.54093847e+01, 1.89411759e+01,
1.56224186e+00, 4.54093847e+01, 2.43529404e+01, 1.56224186e+00, 4.54093847e+01,
5.41176453e+00, 6.24896742e+00, 4.54093847e+01, 1.08235291e+01, 6.24896742e+00,
4.54093847e+01, 1.62352936e+01, 6.24896742e+00, 4.54093847e+01, 2.16470582e+01,
6.24896742e+00, 4.54093847e+01, 2.70588227e+01, 6.24896742e+00, 4.54093847e+01,
8.11764682e+00, 1.09356930e+01, 4.54093847e+01, 1.35294114e+01, 1.09356930e+01,
4.54093847e+01, 1.89411759e+01, 1.09356930e+01, 4.54093847e+01, 2.43529404e+01,
1.09356930e+01, 4.54093847e+01, 2.97647050e+01, 1.09356930e+01, 4.54093847e+01,
1.08235291e+01, 1.56224186e+01, 4.54093847e+01, 1.62352936e+01, 1.56224186e+01,
4.54093847e+01, 2.16470582e+01, 1.56224186e+01, 4.54093847e+01, 2.70588227e+01,
1.56224186e+01, 4.54093847e+01, 3.24705872e+01, 1.56224186e+01, 4.54093847e+01,
1.35294114e+01, 2.03091441e+01, 4.54093847e+01, 1.89411759e+01, 2.03091441e+01,
4.54093847e+01, 2.43529404e+01, 2.03091441e+01, 4.54093847e+01, 2.97647050e+01,
2.03091441e+01, 4.54093847e+01, 3.51764695e+01, 2.03091441e+01, 4.54093847e+01,
-0.00000000e+00, 3.12448372e+00, 4.98280720e+01, 5.41176453e+00, 3.12448372e+00,
4.98280720e+01, 1.08235291e+01, 3.12448372e+00, 4.98280720e+01, 1.62352936e+01,
3.12448372e+00, 4.98280720e+01, 2.16470582e+01, 3.12448372e+00, 4.98280720e+01,
2.70588227e+00, 7.81120928e+00, 4.98280720e+01, 8.11764682e+00, 7.81120928e+00,
4.98280720e+01, 1.35294114e+01, 7.81120928e+00, 4.98280720e+01, 1.89411759e+01,
7.81120928e+00, 4.98280720e+01, 2.43529404e+01, 7.81120928e+00, 4.98280720e+01,
5.41176453e+00, 1.24979349e+01, 4.98280720e+01, 1.08235291e+01, 1.24979349e+01,
4.98280720e+01, 1.62352936e+01, 1.24979349e+01, 4.98280720e+01, 2.16470582e+01,
1.24979349e+01, 4.98280720e+01, 2.70588227e+01, 1.24979349e+01, 4.98280720e+01,
8.11764682e+00, 1.71846604e+01, 4.98280720e+01, 1.35294114e+01, 1.71846604e+01,
4.98280720e+01, 1.89411759e+01, 1.71846604e+01, 4.98280720e+01, 2.43529404e+01,
1.71846604e+01, 4.98280720e+01, 2.97647050e+01, 1.71846604e+01, 4.98280720e+01,
1.08235291e+01, 2.18713860e+01, 4.98280720e+01, 1.62352936e+01, 2.18713860e+01,
4.98280720e+01, 2.16470582e+01, 2.18713860e+01, 4.98280720e+01, 2.70588227e+01,
2.18713860e+01, 4.98280720e+01, 3.24705872e+01, 2.18713860e+01, 4.98280720e+01,
0.00000000e+00, 0.00000000e+00, 5.42467592e+01, 5.41176453e+00, 0.00000000e+00,
5.42467592e+01, 1.08235291e+01, 0.00000000e+00, 5.42467592e+01, 1.62352936e+01,
0.00000000e+00, 5.42467592e+01, 2.16470582e+01, 0.00000000e+00, 5.42467592e+01,
2.70588227e+00, 4.68672556e+00, 5.42467592e+01, 8.11764682e+00, 4.68672556e+00,
5.42467592e+01, 1.35294114e+01, 4.68672556e+00, 5.42467592e+01, 1.89411759e+01,
4.68672556e+00, 5.42467592e+01, 2.43529404e+01, 4.68672556e+00, 5.42467592e+01,
5.41176453e+00, 9.37345114e+00, 5.42467592e+01, 1.08235291e+01, 9.37345114e+00,
5.42467592e+01, 1.62352936e+01, 9.37345114e+00, 5.42467592e+01, 2.16470582e+01,
9.37345114e+00, 5.42467592e+01, 2.70588227e+01, 9.37345114e+00, 5.42467592e+01,
8.11764682e+00, 1.40601767e+01, 5.42467592e+01, 1.35294114e+01, 1.40601767e+01,
5.42467592e+01, 1.89411759e+01, 1.40601767e+01, 5.42467592e+01, 2.43529404e+01,
1.40601767e+01, 5.42467592e+01, 2.97647050e+01, 1.40601767e+01, 5.42467592e+01,
1.08235291e+01, 1.87469023e+01, 5.42467592e+01, 1.62352936e+01, 1.87469023e+01,
5.42467592e+01, 2.16470582e+01, 1.87469023e+01, 5.42467592e+01, 2.70588227e+01,
1.87469023e+01, 5.42467592e+01, 3.24705872e+01, 1.87469023e+01, 5.42467592e+01,
0.00000000e+00, 0.00000000e+00, 5.80262115e+01, -3.02356180e+00, 0.00000000e+00,
5.80262115e+01, 9.44863063e+00, 0.00000000e+00, 5.80262115e+01, 6.42506883e+00,
0.00000000e+00, 5.80262115e+01, 1.88972613e+01, 0.00000000e+00, 5.80262115e+01,
1.58736995e+01, 0.00000000e+00, 5.80262115e+01, 9.44863063e+00, 9.44863063e+00,
5.80262115e+01, 6.42506883e+00, 9.44863063e+00, 5.80262115e+01, 1.88972613e+01,
9.44863063e+00, 5.80262115e+01, 1.58736995e+01, 9.44863063e+00, 5.80262115e+01,
2.83458919e+01, 9.44863063e+00, 5.80262115e+01, 2.53223301e+01, 9.44863063e+00,
5.80262115e+01, 1.88972613e+01, 1.88972613e+01, 5.80262115e+01, 1.58736995e+01,
1.88972613e+01, 5.80262115e+01, 2.83458919e+01, 1.88972613e+01, 5.80262115e+01,
2.53223301e+01, 1.88972613e+01, 5.80262115e+01 ]</q>
<p shape='(1, 573)'>[ 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00 ]</p>
<m shape='(191)'>[ 4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 5.11967350e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.43053050e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 5.11967350e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04,
4.43053050e+04, 4.91843353e+04, 4.91843353e+04, 4.91843353e+04, 5.11967350e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04, 2.91651223e+04,
2.91651223e+04 ]</m>
<names shape='(191)'>[ Mg, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Si, Al, Mg, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Si,
Al, Al, Al, Al, Al,
Al, Al, Al, Mg, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Si, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Si, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Mg, Al, Al,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Si, Al, Al, Al,
Al, Al, Al, Al, Si,
Al, Al, Al, Al, Al,
Al, Al, Al, Al, Al,
Al, Mg, Al, Al, Al,
Al, Al, Al, Al, Al,
Mg, Al, Al, Al, Si,
O, O, O, O, O,
O, O, O, O, O,
O, O, O, O, O,
O ]</names>
</beads>
<cell shape='(3, 3)'>[ 2.70588227e+01, 1.35294114e+01, 0.00000000e+00, 0.00000000e+00, 2.34336279e+01,
0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 7.31440205e+01 ]</cell>
<initialize nbeads='1'>
<velocities mode='thermal' units='ase'>800</velocities>
</initialize>
<ensemble>
<temperature units='ase'>800</temperature>
</ensemble>
<forces>
<force forcefield='pet-mad'>
</force>
</forces>
<motion mode='multi'>
<motion mode='dynamics'>
<dynamics mode='nvt'>
<timestep units='ase'>0.4911347394232032</timestep>
<thermostat mode='svr'>
<tau units='ase'>4.911347394232032</tau>
</thermostat>
</dynamics>
</motion>
<motion mode='atomswap'>
<atomswap>
<nxc>0.1</nxc>
<names>[ Al, Si, Mg, O]</names>
</atomswap>
</motion>
</motion>
</system>
</simulation>
--- end input file content ---
@system.bind: Binding the forces
@initializer: Initializer (stage 2) parsing velocities object.
@simulation.run: Average timings at MD step 0. t/step: 1.37616e+00
@simulation.run: Average timings at MD step 1. t/step: 6.26788e-01
@simulation.run: Average timings at MD step 2. t/step: 3.68626e-01
@simulation.run: Average timings at MD step 3. t/step: 4.10013e-01
@simulation.run: Average timings at MD step 4. t/step: 3.58863e-01
@simulation.run: Average timings at MD step 5. t/step: 3.83273e-01
@simulation.run: Average timings at MD step 6. t/step: 3.45273e-01
@simulation.run: Average timings at MD step 7. t/step: 4.14647e-01
@simulation.run: Average timings at MD step 8. t/step: 3.61163e-01
@simulation.run: Average timings at MD step 9. t/step: 4.27952e-01
@simulation.run: Average timings at MD step 10. t/step: 3.66755e-01
@simulation.run: Average timings at MD step 11. t/step: 4.19497e-01
@simulation.run: Average timings at MD step 12. t/step: 7.25109e-01
@simulation.run: Average timings at MD step 13. t/step: 3.99104e-01
@simulation.run: Average timings at MD step 14. t/step: 4.16743e-01
@simulation.run: Average timings at MD step 15. t/step: 4.21087e-01
@simulation.run: Average timings at MD step 16. t/step: 3.65427e-01
@simulation.run: Average timings at MD step 17. t/step: 4.29073e-01
@simulation.run: Average timings at MD step 18. t/step: 3.77333e-01
@simulation.run: Average timings at MD step 19. t/step: 4.19869e-01
@simulation.run: Average timings at MD step 20. t/step: 3.94575e-01
@simulation.run: Average timings at MD step 21. t/step: 4.88821e-01
@simulation.run: Average timings at MD step 22. t/step: 3.87254e-01
@simulation.run: Average timings at MD step 23. t/step: 4.21123e-01
@simulation.run: Average timings at MD step 24. t/step: 3.78220e-01
@simulation.run: Average timings at MD step 25. t/step: 4.97568e-01
@simulation.run: Average timings at MD step 26. t/step: 3.92988e-01
@simulation.run: Average timings at MD step 27. t/step: 4.75909e-01
@simulation.run: Average timings at MD step 28. t/step: 3.85914e-01
@simulation.run: Average timings at MD step 29. t/step: 4.95407e-01
@simulation.run: Average timings at MD step 30. t/step: 3.90073e-01
@simulation.run: Average timings at MD step 31. t/step: 4.76712e-01
@simulation.run: Average timings at MD step 32. t/step: 3.81382e-01
@simulation.run: Average timings at MD step 33. t/step: 4.92934e-01
@simulation.run: Average timings at MD step 34. t/step: 4.06611e-01
@simulation.run: Average timings at MD step 35. t/step: 4.53342e-01
@simulation.run: Average timings at MD step 36. t/step: 3.83616e-01
@simulation.run: Average timings at MD step 37. t/step: 4.89239e-01
@simulation.run: Average timings at MD step 38. t/step: 3.87391e-01
@simulation.run: Average timings at MD step 39. t/step: 4.23603e-01
@simulation.run: Average timings at MD step 40. t/step: 3.79894e-01
@simulation.run: Average timings at MD step 41. t/step: 4.85070e-01
@simulation.run: Average timings at MD step 42. t/step: 3.86276e-01
@simulation.run: Average timings at MD step 43. t/step: 4.72796e-01
@simulation.run: Average timings at MD step 44. t/step: 3.81214e-01
@simulation.run: Average timings at MD step 45. t/step: 4.37862e-01
@simulation.run: Average timings at MD step 46. t/step: 3.69235e-01
@simulation.run: Average timings at MD step 47. t/step: 4.61076e-01
@simulation.run: Average timings at MD step 48. t/step: 3.87221e-01
@simulation.run: Average timings at MD step 49. t/step: 1.25928e+00
@simulation.run: Average timings at MD step 50. t/step: 4.23959e-01
@simulation.run: Average timings at MD step 51. t/step: 4.20723e-01
@simulation.run: Average timings at MD step 52. t/step: 3.92343e-01
@simulation.run: Average timings at MD step 53. t/step: 4.39362e-01
@simulation.run: Average timings at MD step 54. t/step: 3.86080e-01
@simulation.run: Average timings at MD step 55. t/step: 4.20244e-01
@simulation.run: Average timings at MD step 56. t/step: 3.85428e-01
@simulation.run: Average timings at MD step 57. t/step: 4.32564e-01
@simulation.run: Average timings at MD step 58. t/step: 7.78606e-01
@simulation.run: Average timings at MD step 59. t/step: 4.39392e-01
@simulation.run: Average timings at MD step 60. t/step: 3.85344e-01
@simulation.run: Average timings at MD step 61. t/step: 4.24933e-01
@simulation.run: Average timings at MD step 62. t/step: 3.88027e-01
@simulation.run: Average timings at MD step 63. t/step: 4.37152e-01
@simulation.run: Average timings at MD step 64. t/step: 3.81072e-01
@simulation.run: Average timings at MD step 65. t/step: 4.56959e-01
@simulation.run: Average timings at MD step 66. t/step: 3.75730e-01
@simulation.run: Average timings at MD step 67. t/step: 4.79268e-01
@simulation.run: Average timings at MD step 68. t/step: 7.60598e-01
@simulation.run: Average timings at MD step 69. t/step: 4.98282e-01
@simulation.run: Average timings at MD step 70. t/step: 3.83121e-01
@simulation.run: Average timings at MD step 71. t/step: 4.31491e-01
@simulation.run: Average timings at MD step 72. t/step: 3.88567e-01
@simulation.run: Average timings at MD step 73. t/step: 4.56474e-01
@simulation.run: Average timings at MD step 74. t/step: 4.13692e-01
@simulation.run: Average timings at MD step 75. t/step: 4.60695e-01
@simulation.run: Average timings at MD step 76. t/step: 4.02555e-01
@simulation.run: Average timings at MD step 77. t/step: 4.25380e-01
@simulation.run: Average timings at MD step 78. t/step: 7.58412e-01
@simulation.run: Average timings at MD step 79. t/step: 4.43541e-01
The simulation generates output files that can be parsed and visualized from Python.
data, info = read_output("nvt_atomxc.out")
trj = read_trajectory("nvt_atomxc.pos_0.xyz")
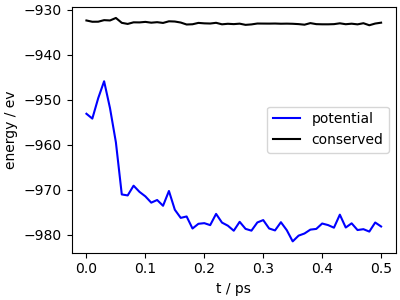
fig, ax = plt.subplots(1, 1, figsize=(4, 3), constrained_layout=True)
ax.plot(data["time"], data["potential"], "b-", label="potential")
ax.plot(data["time"], data["conserved"] - 4, "k-", label="conserved")
ax.set_xlabel("t / ps")
ax.set_ylabel("energy / ev")
ax.legend()
@process_units: Interpreting input with dimension length, units angstrom and cell units angstrom
@process_units: Interpreting input with dimension length, units angstrom and cell units angstrom
@process_units: Interpreting input with dimension length, units angstrom and cell units angstrom
@process_units: Interpreting input with dimension length, units angstrom and cell units angstrom
@process_units: Interpreting input with dimension length, units angstrom and cell units angstrom
<matplotlib.legend.Legend object at 0x7f059423f200>
The trajectory (which is started from oxygen molecules placed on top of the surface) shows quick relaxation to an oxide layer. If you look carefully, you’ll also see that Mg and Si atoms tend to cluster together, and accumulate at the surface.
chemiscope.show(
structures=trj,
properties={
"time": data["time"][::10],
"potential": data["potential"][::10],
"temperature": data["temperature"][::10],
},
mode="default",
settings=chemiscope.quick_settings(
map_settings={
"x": {"property": "time", "scale": "linear"},
"y": {"property": "potential", "scale": "linear"},
},
structure_settings={
"unitCell": True,
},
trajectory=True,
),
)
Molecular dynamics with LAMMPS¶
We now run the same MD with LAMMPS. To run a LAMMPS
calculation with a metatomic potential, one needs a LAMMPS build that contains an
appropriate pair style. You can compile it from source, or fetch it from the metatensor channel on
conda. One can then just include in the input a pair_style metatomic that points
to the exported model and a single pair_coeff command that specifies the mapping
from LAMMPS types to the atomic types the model can handle. The first two arguments
must be * * so as to span all LAMMPS atom types. This is followed by a list of N
arguments that specify the mapping of metatomic atomic types to LAMMPS types, where N
is the number of LAMMPS atom types.
with open("data/al6xxx-o2.in", "r") as f:
print(f.read())
units metal # Angstroms, eV, picoseconds
atom_style atomic
read_data al6xxx-o2.data
# loads pet-mad-model
pair_style metatomic &
pet-mad-s-v1.0.2.pt &
device cpu &
extensions extensions/
# define interactions between all atoms and maps the LAMMPS types to elements
pair_coeff * * 13 12 8 14
# allow for large neighbor list to accommodate large adaptive cutoff
neighbor 2.0 bin
neigh_modify one 100000 page 1000000 binsize 5.5
timestep 0.005
dump myDump all xyz 10 trajectory.xyz
dump_modify myDump element Al Mg O Si
thermo_style multi
thermo 1
velocity all create 800 87287 mom yes rot yes
fix 1 all nvt temp 800 800 0.10
# fix 2 all atom/swap 1 1 12345 800 types 1 2
# fix 2 all atom/swap 1 1 12345 800 types 1 3
# fix 2 all atom/swap 1 1 12345 800 types 1 4
run 80
Warning
Be aware that the extensions are compiled files and depend on your operating system.
Usually you have re-export the extensions for different systems! You can do this
by running the appropriate parts of this file, or using the mtt export
command-line utility.
We also save the geometry to a LAMMPS data file and finally run the simulation.
ase.io.write("al6xxx-o2.data", al_surface, format="lammps-data", masses=True)
subprocess.check_call(["lmp", "-in", "data/al6xxx-o2.in"])
0
The resulting trajectory is qualitatively consistent with what
we observed with i-PI.
lmp_trj = ase.io.read("trajectory.xyz", ":")
chemiscope.show(structures=lmp_trj, mode="structure")
Total running time of the script: (2 minutes 7.083 seconds)