metatensor¶
Metatensor
is a library providing a cross-platform data interchange API
for atomistic simulation and beyond. It also powers
metatomic – an API to define atomistic models that can be
used to run simulations using several different atomistic
simulation packages and metatrain a set of tools to facilitate
training and evaluating ML models.
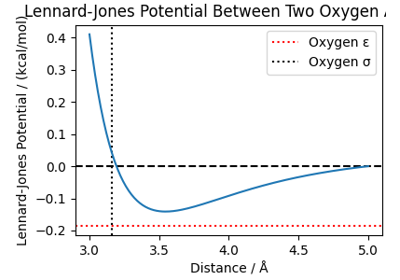
Atomistic Water Model for Molecular Dynamics
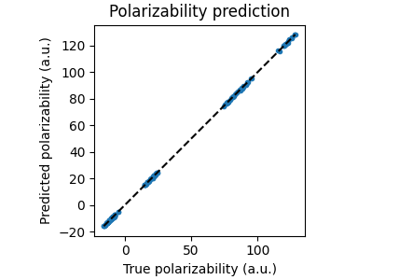
Equivariant linear model for polarizability
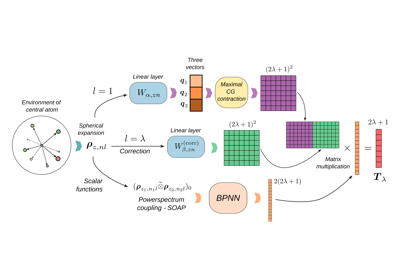
Equivariant model for tensorial properties based on scalar features
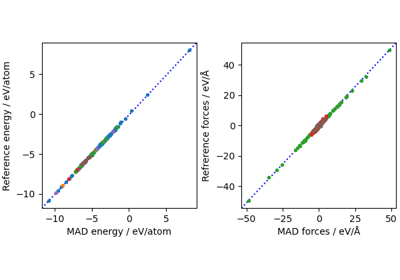
The PET-MAD universal potential
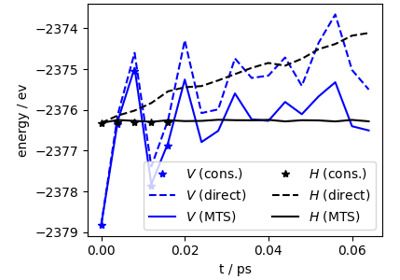
MD using direct-force predictions with PET-MAD
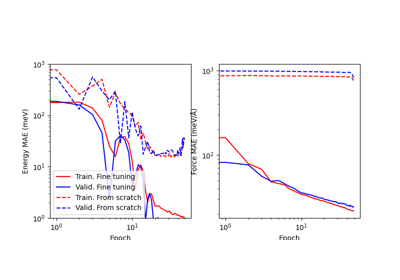
Fine-tuning the PET-MAD universal potential
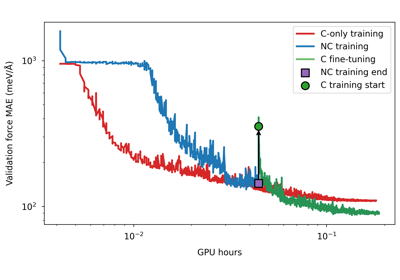
Conservative fine-tuning for a PET model
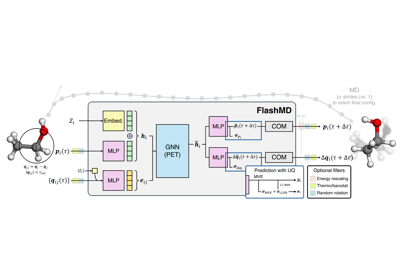
Long-stride trajectories with a universal FlashMD model
Computing NMR shielding tensors using ShiftML
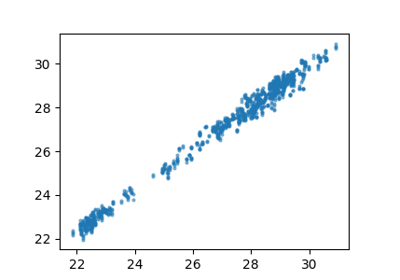
Hamiltonian Learning for Molecules with Indirect Targets
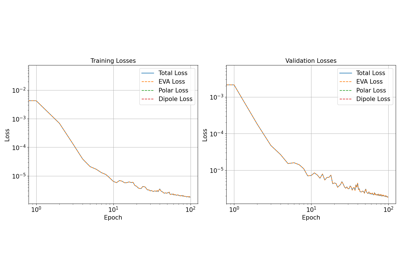
ML collective variables in PLUMED with metatomic
