Note
Go to the end to download the full example code.
Computing NMR shielding tensors using ShiftML¶
- Authors:
Michele Ceriotti @ceriottm
This example shows how to compute NMR shielding tensors using a point-edge transformer model trained on the ShiftML dataset.
import os
import zipfile
import chemiscope
import matplotlib.pyplot as plt
import numpy as np
import requests
from ase.io import read
from shiftml.ase import ShiftML
Create a ShiftML calculator and fetch a dataset¶
calculator = ShiftML("ShiftML3")
filename = "ShiftML_poly.zip"
if not os.path.exists(filename):
url = (
"https://archive.materialscloud.org/records/j2fka-sda13/files/ShiftML_poly.zip"
)
response = requests.get(url)
response.raise_for_status()
with open(filename, "wb") as f:
f.write(response.content)
with zipfile.ZipFile(filename, "r") as zip_ref:
for file in ["ShiftML_poly/Cocaine/cocaine_QuantumEspresso.xyz"]:
target = os.path.basename(file)
with zip_ref.open(file) as source, open(target, "wb") as dest:
dest.write(source.read())
frames_cocaine = read("cocaine_QuantumEspresso.xyz", index=":16")
reference = [frame.arrays["CS"] for frame in frames_cocaine]
2026-02-17 16:26:47,765 - INFO - Found model version in url_resolve
2026-02-17 16:26:47,765 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_0.pt?download=1
2026-02-17 16:26:47,765 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:49,508 - INFO - Downloaded ShiftML30 and saved to /home/runner/.cache/shiftml/ShiftML30
2026-02-17 16:26:49,670 - INFO - Found model version in url_resolve
2026-02-17 16:26:49,670 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_1.pt?download=1
2026-02-17 16:26:49,670 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:51,232 - INFO - Downloaded ShiftML31 and saved to /home/runner/.cache/shiftml/ShiftML31
2026-02-17 16:26:51,385 - INFO - Found model version in url_resolve
2026-02-17 16:26:51,386 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_2.pt?download=1
2026-02-17 16:26:51,386 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:52,884 - INFO - Downloaded ShiftML32 and saved to /home/runner/.cache/shiftml/ShiftML32
2026-02-17 16:26:53,037 - INFO - Found model version in url_resolve
2026-02-17 16:26:53,037 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_3.pt?download=1
2026-02-17 16:26:53,037 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:54,646 - INFO - Downloaded ShiftML33 and saved to /home/runner/.cache/shiftml/ShiftML33
2026-02-17 16:26:54,799 - INFO - Found model version in url_resolve
2026-02-17 16:26:54,799 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_4.pt?download=1
2026-02-17 16:26:54,800 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:56,349 - INFO - Downloaded ShiftML34 and saved to /home/runner/.cache/shiftml/ShiftML34
2026-02-17 16:26:56,505 - INFO - Found model version in url_resolve
2026-02-17 16:26:56,505 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_5.pt?download=1
2026-02-17 16:26:56,505 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:58,148 - INFO - Downloaded ShiftML35 and saved to /home/runner/.cache/shiftml/ShiftML35
2026-02-17 16:26:58,302 - INFO - Found model version in url_resolve
2026-02-17 16:26:58,302 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_6.pt?download=1
2026-02-17 16:26:58,303 - INFO - Model not found in cache, downloading it
2026-02-17 16:26:59,821 - INFO - Downloaded ShiftML36 and saved to /home/runner/.cache/shiftml/ShiftML36
2026-02-17 16:26:59,975 - INFO - Found model version in url_resolve
2026-02-17 16:26:59,975 - INFO - Resolving model version to model files at url: https://zenodo.org/records/15767390/files/model_7.pt?download=1
2026-02-17 16:26:59,976 - INFO - Model not found in cache, downloading it
2026-02-17 16:27:01,561 - INFO - Downloaded ShiftML37 and saved to /home/runner/.cache/shiftml/ShiftML37
Predicts isotropic chemical shielding tensors, including uncertainty¶
predicted = [calculator.get_cs_iso_ensemble(frame) for frame in frames_cocaine]
Make a plot for all the H shielding values
h_shieldings = np.hstack(
[
[
r[f.symbols == "H"],
p[f.symbols == "H"].mean(axis=1),
p[f.symbols == "H"].std(axis=1),
]
for r, p, f in zip(reference, predicted, frames_cocaine)
]
)
and of the uncertainty
fig, ax = plt.subplots(figsize=(4, 3))
ax.plot(h_shieldings[0], h_shieldings[1], "o", markersize=2, alpha=0.5)
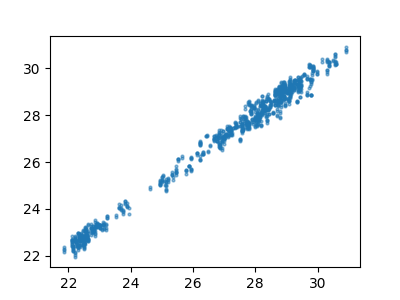
[<matplotlib.lines.Line2D object at 0x7fa61262e510>]
fig, ax = plt.subplots(figsize=(4, 3))
ax.loglog(
h_shieldings[2],
np.abs(h_shieldings[0] - h_shieldings[1]),
"o",
markersize=2,
alpha=0.5,
)
ax.plot([0.2, 0.8], [0.2, 0.8], "k--", lw=0.5)
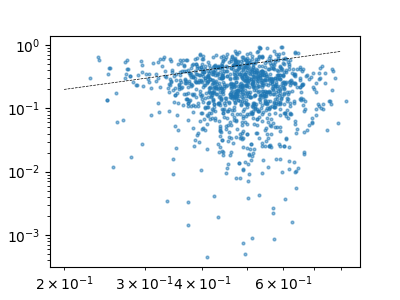
[<matplotlib.lines.Line2D object at 0x7fa61292c4d0>]
Anisotropic shielding tensors¶
tensors = [calculator.get_cs_tensor(frame) for frame in frames_cocaine]
h_tensors = np.array(
[
(t[iat] if f.symbols[iat] == "H" else np.zeros((3, 3)))
for t, f in zip(tensors, frames_cocaine)
for iat in range(len(f))
]
)
chemiscope.show(
frames_cocaine,
shapes={
"cs_ellipsoid": {
"kind": "ellipsoid",
"parameters": {
"global": {},
"atom": [
chemiscope.ellipsoid_from_tensor(cs_h * 0.05) for cs_h in h_tensors
],
},
}
},
mode="structure",
settings=chemiscope.quick_settings(
periodic=True,
structure_settings={
"shape": ["cs_ellipsoid"],
},
),
)
Total running time of the script: (1 minutes 58.991 seconds)