Note
Go to the end to download the full example code.
Equivariant linear model for polarizability¶
- Authors:
Paolo Pegolo @ppegolo
In this example, we demonstrate how to construct a metatensor atomistic model for the polarizability tensor
of molecular systems. This example uses the featomic library to compute
equivariant descriptors, and scikit-learn to train a linear regression model.
The model can then be used in an ASE calculator.
You could also have a look at
this recipe based on scalar/tensorial models,
which provides an alternative approach for equivariant learning
of tensors.
from typing import Dict, List, Optional
import ase.io
import chemiscope
# Simulation and visualization tools
import matplotlib.pyplot as plt
import metatensor.torch as mts
# Model wrapping and execution tools
import numpy as np
import torch
from featomic.torch import SphericalExpansion
from featomic.torch.clebsch_gordan import (
EquivariantPowerSpectrum,
cartesian_to_spherical,
)
from metatensor.torch import Labels, TensorBlock, TensorMap
from metatensor.torch.learn.nn import EquivariantLinear
from metatomic.torch import (
AtomisticModel,
ModelCapabilities,
ModelEvaluationOptions,
ModelMetadata,
ModelOutput,
System,
load_atomistic_model,
systems_to_torch,
)
from metatomic.torch.ase_calculator import MetatomicCalculator
# Core libraries
from sklearn.linear_model import RidgeCV
torch.set_default_dtype(torch.float64) # FIXME: This is a temporary fix
Polarizability tensor¶
The polarizability tensor describes the response of a molecule to an external electric field.
It is a rank-2 symmetric tensor and it can be decomposed into irreducible spherical components. Due to its symmetry, only the components with \(\lambda=0\) and \(\lambda=2\) are non-zero. The \(\lambda=0\) component is a scalar, while the \(\lambda=2\) component corresponds to a symmetric traceless matrix
Load the training data¶
We load a simple dataset of \(\mathrm{C}_5\mathrm{NH}_7\) molecules and their polarizability tensors stored in extended XYZ format. We also visualize the polarizability as ellipsoids to demonstrate the anisotropy of this molecular property.
ase_frames = ase.io.read("data/qm7x_reduced_100.xyz", index=":")
ellipsoids = chemiscope.ase_tensors_to_ellipsoids(
ase_frames, "polarizability", scale=0.15
)
ellipsoids["parameters"]["global"]["color"] = "#FF8800"
cs = chemiscope.show(
ase_frames,
shapes={"alpha": ellipsoids},
mode="structure",
settings=chemiscope.quick_settings(
structure_settings={"shape": ["alpha"]}, trajectory=True
),
)
cs
Read the polarizability tensors and store them in a TensorMap¶
We extract the polarizability tensors from the extended XYZ file and store them in a
metatensor.torch.TensorMap. The polarizability tensors are stored as
Cartesian tensors.
cartesian_polarizability = np.stack(
[frame.info["polarizability"].reshape(3, 3) for frame in ase_frames]
)
cartesian_tensormap = TensorMap(
keys=Labels.single(),
blocks=[
TensorBlock(
samples=Labels.range("system", len(ase_frames)),
components=[Labels.range(name, 3) for name in ["xyz_1", "xyz_2"]],
properties=Labels(["polarizability"], torch.tensor([[0]])),
values=torch.from_numpy(cartesian_polarizability).unsqueeze(-1),
)
],
)
print(cartesian_tensormap)
print(cartesian_tensormap.block(0))
TensorMap with 1 blocks
keys: _
0
TensorBlock
samples (100): ['system']
components (3, 3): ['xyz_1', 'xyz_2']
properties (1): ['polarizability']
gradients: None
We also convert the Cartesian tensors to irreducible spherical
tensors using the
featomic.torch.clebsch_gordan.cartesian_to_spherical()
function. Now the polarizability is stored in a
metatensor.torch.TensorMap object
that contains two metatensor.torch.TensorBlock labeled by
the irreducible spherical components as keys.
spherical_tensormap = mts.remove_dimension(
cartesian_to_spherical(cartesian_tensormap, components=["xyz_1", "xyz_2"]),
"keys",
"_",
)
print(spherical_tensormap)
print(spherical_tensormap.block(0))
print(spherical_tensormap.block(1))
TensorMap with 2 blocks
keys: o3_lambda o3_sigma
0 1
2 1
TensorBlock
samples (100): ['system']
components (1): ['o3_mu']
properties (1): ['polarizability']
gradients: None
TensorBlock
samples (100): ['system']
components (5): ['o3_mu']
properties (1): ['polarizability']
gradients: None
Split the dataset¶
The dataset is split into training and test sets using a 80/20 ratio. We also visualize the polarizability as ellipsoids to e into training and test sets according to the indices previously defined.
n_frames = len(ase_frames)
train_idx = np.random.choice(n_frames, int(0.8 * n_frames), replace=False)
test_idx = np.setdiff1d(np.arange(n_frames), train_idx)
spherical_tensormap_train = mts.slice(
spherical_tensormap,
"samples",
Labels("system", torch.from_numpy(train_idx).reshape(-1, 1)),
)
cartesian_tensormap_test = mts.slice(
cartesian_tensormap,
"samples",
Labels("system", torch.from_numpy(test_idx).reshape(-1, 1)),
)
Equivariant model for polarizability¶
The polarizability tensor can be predicted using equivariant linear models. In this
example, we use the featomic library to compute equivariant \(\lambda\)-SOAP
descriptors, adapted to the symmetry of the irreducible components of
\(\boldsymbol{\alpha}\) and scikit-learn to train a symmetry-adapted
linear ridge regression model.
Utility functions¶
We define a couple of utility functions to convert the spherical polarizability tensor back to a Cartesian tensor.
The first utility function takes the \(\lambda=2\) spherical components and reconstructs the \(3 \times 3\) symmetric traceless matrix.
def matrix_from_l2_components(l2: torch.Tensor) -> torch.Tensor:
"""
Inverts the spherical projection function for a symmetric, traceless 3x3 tensor.
Given:
l2 : array-like of 5 components
These are the irreducible spherical components multiplied by sqrt(2),
i.e., l2 = sqrt(2) * [t0, t1, t2, t3, t4], where:
t0 = (A[0,1] + A[1,0])/2
t1 = (A[1,2] + A[2,1])/2
t2 = (2*A[2,2] - A[0,0] - A[1,1])/(2*sqrt(3))
t3 = (A[0,2] + A[2,0])/2
t4 = (A[0,0] - A[1,1])/2
Returns:
A : (3,3) numpy array
The symmetric, traceless matrix reconstructed from the components.
"""
# Recover the t_i by dividing by sqrt(2)
sqrt2 = torch.sqrt(torch.tensor(2.0, dtype=l2.dtype))
sqrt3 = torch.sqrt(torch.tensor(3.0, dtype=l2.dtype))
l2 = l2 / sqrt2
# Allocate the 3x3 matrix A
A = torch.empty((3, 3), dtype=l2.dtype)
# Diagonal entries:
# Use the traceless condition A[0,0] + A[1,1] + A[2,2] = 0.
# Also, from the definitions:
# t4 = (A[0,0] - A[1,1]) / 2
# t2 = (2*A[2,2] - A[0,0] - A[1,1])/(2*sqrt3)
#
# Solve for A[0,0] and A[1,1]:
A[0, 0] = -(sqrt3 * l2[2]) / 3 + l2[4]
A[1, 1] = -(sqrt3 * l2[2]) / 3 - l2[4]
A[2, 2] = (2 * sqrt3 * l2[2]) / 3 # Since A[2,2] = - (A[0,0] + A[1,1])
# Off-diagonals:
A[0, 1] = l2[0]
A[1, 0] = l2[0]
A[1, 2] = l2[1]
A[2, 1] = l2[1]
A[0, 2] = l2[3]
A[2, 0] = l2[3]
return A
The second utility function takes the spherical polarizability tensor and uses the
matrix_from_l2_components function to reconstruct symmetric traceless part of the
Cartesian tensor, and then combines it with the scalar part to reconstruct the full
Cartesian tensor.
def spherical_to_cartesian(spherical_tensor: TensorMap) -> TensorMap:
dtype = spherical_tensor[0].values.dtype
new_block: Dict[int, torch.Tensor] = {}
eye = torch.eye(3, dtype=dtype)
sqrt3 = torch.sqrt(torch.tensor(3.0, dtype=dtype))
block_0 = spherical_tensor[0]
block_2 = spherical_tensor[1]
system_ids = block_0.samples.values.flatten()
for i, A in enumerate(system_ids):
new_block[int(A)] = -block_0.values[
i
].flatten() * eye / sqrt3 + matrix_from_l2_components(
block_2.values[i].squeeze()
)
return TensorMap(
keys=Labels(["_"], torch.tensor([[0]], dtype=torch.int32)),
blocks=[
TensorBlock(
samples=Labels(
"system",
torch.tensor(
[k for k in new_block.keys()], dtype=torch.int32
).reshape(-1, 1),
),
components=[
Labels(
"xyz_1",
torch.tensor([0, 1, 2], dtype=torch.int32).reshape(-1, 1),
),
Labels(
"xyz_2",
torch.tensor([0, 1, 2], dtype=torch.int32).reshape(-1, 1),
),
],
properties=Labels(["_"], torch.tensor([[0]], dtype=torch.int32)),
values=torch.stack(
[new_block[A] for A in new_block]
).unsqueeze( # .roll((-1, -1), dims=(1, 2))
-1
),
)
],
)
Implementation of a torch module for polarizability¶
In order to implement a polarizability model compatible with metatomic, we need to
we need to define a torch.nn.Module with a forward method that takes a list of
metatomic.torch.System and returns a dictionary of
metatensor.torch.TensorMap objects. The forward method must be compatible
with TorchScript.
Here, the PolarizabilityModel class is defined. It takes as input a
SphericalExpansion object, a list of atomic types, and dtype. The
forward method computes the descriptors and applies the linear model to predict
the polarizability tensor.
class PolarizabilityModel(torch.nn.Module):
def __init__(
self,
spex_calculator: SphericalExpansion,
atomic_types: List[int],
dtype: torch.dtype = None,
) -> None:
super().__init__()
if dtype is None:
dtype = torch.float64
self.dtype = dtype
device = torch.device("cpu")
self.hypers = spex_calculator.parameters
# Check that the atomic types are unique
assert len(set(atomic_types)) == len(atomic_types)
self.atomic_types = atomic_types
# Define lambda soap calculator
self.lambda_soap_calculator = EquivariantPowerSpectrum(
spex_calculator, dtype=self.dtype
)
self.selected_keys = mts.Labels(
["o3_lambda", "o3_sigma"], torch.tensor([[0, 1], [2, 1]])
)
self._compute_metadata()
# Define the linear model that wraps the ridge regression results
in_keys = self.metadata.keys
in_features = [block.values.shape[-1] for block in self.metadata]
out_features = [1 for _ in self.metadata]
out_properties = [mts.Labels.single() for _ in self.metadata]
self.linear_model = EquivariantLinear(
in_keys=in_keys,
in_features=in_features,
out_features=out_features,
out_properties=out_properties,
dtype=self.dtype,
device=device,
)
def fit(self, training_systems, training_targets, alphas=None):
lambda_soap = self._compute_descriptor(training_systems)
ridges: List[RidgeCV] = []
for k in lambda_soap.keys:
X = lambda_soap.block(k).values
y = training_targets.block(k).values
n_samples, n_components, n_properties = X.shape
X = X.reshape(n_samples * n_components, n_properties)
y = y.reshape(n_samples * n_components, -1)
ridge = RidgeCV(alphas=alphas, fit_intercept=int(k["o3_lambda"]) == 0)
ridge.fit(X, y)
ridges.append(ridge)
self._apply_weights(ridges)
def _compute_metadata(self):
# Create dummy system to get the property dimension
dummy_system = systems_to_torch(
ase.Atoms(
numbers=self.atomic_types,
positions=[[i / 4, 0, 0] for i in range(len(self.atomic_types))],
)
)
self.metadata = self.lambda_soap_calculator.compute_metadata(
dummy_system, selected_keys=self.selected_keys, neighbors_to_properties=True
).keys_to_samples("center_type")
def _apply_weights(self, ridges: List[RidgeCV]) -> None:
with torch.no_grad():
for model, ridge in zip(self.linear_model.module_map, ridges):
model.weight.copy_(torch.tensor(ridge.coef_, dtype=self.dtype))
if model.bias is not None:
model.bias.copy_(torch.tensor(ridge.intercept_, dtype=self.dtype))
def _compute_descriptor(self, systems: List[System]) -> TensorMap:
# Actually compute lambda-SOAP power spectrum
lambda_soap = self.lambda_soap_calculator(
systems,
selected_keys=self.selected_keys,
neighbors_to_properties=True,
)
# Move the `center_type` keys to the sample dimension
lambda_soap = lambda_soap.keys_to_samples("center_type")
# Polarizability is a "per-structure" quantity. We don't need to keep
# `center_type` and `atom` in samples, as we only need to predict the
# polarizability of the whole structure.
# A simple way to do so, is summing the descriptors over the `center_type` and
# `atom` sample dimensions.
lambda_soap = mts.sum_over_samples(lambda_soap, ["center_type", "atom"])
return lambda_soap
def forward(
self,
systems: List[System], # noqa
outputs: Dict[str, ModelOutput], # noqa
selected_atoms: Optional[Labels] = None, # noqa
) -> Dict[str, TensorMap]: # noqa
if list(outputs.keys()) != ["cookbook::polarizability"]:
raise ValueError(
f"`outputs` keys ({', '.join(outputs.keys())}) contain unsupported "
"keys. Only 'cookbook::polarizability' is supported."
)
if outputs["cookbook::polarizability"].per_atom:
raise NotImplementedError("per-atom polarizabilities are not supported.")
# Compute the descriptors
lambda_soap = self._compute_descriptor(systems)
# Apply the linear model
prediction = self.linear_model(lambda_soap)
prediction = spherical_to_cartesian(prediction)
# fails at saving due to TorchScript
return {"cookbook::polarizability": prediction}
Train the polarizability model¶
We train the polarizability model using the training data. We need to define the
hyperparameters for the featomic.torch.SphericalExpansion calculator and the
atomic types in the dataset to initialize the module.
hypers = {
"cutoff": {"radius": 4.5, "smoothing": {"type": "ShiftedCosine", "width": 0.1}},
"density": {"type": "Gaussian", "width": 0.3},
"basis": {
"type": "TensorProduct",
"max_angular": 3,
"radial": {"type": "Gto", "max_radial": 3},
},
}
atomic_types = np.unique(
np.concatenate([frame.numbers for frame in ase_frames])
).tolist()
spherical_expansion_calculator = SphericalExpansion(**hypers)
Convert ASE frames to metatensor.atomistic.System objects.
Instantiate the polarizability model.
model = PolarizabilityModel(
spherical_expansion_calculator,
atomic_types,
dtype=torch.float64,
)
The ridge regression is performed as with a scikit-learn-style fit method, and
the resulting weights are then used to initialize a
metatensor.learn.nn.EquivariantLinear module.
The PolariabilityModel.fit() method takes the training systems in
metatomic.torch.System format and the training target
metatensor.torch.TensorMap as input. The alphas parameter is used to
specify the range of regularization parameters to be tested.
systems_train = [
system.to(dtype=torch.float64)
for system in systems_to_torch([ase_frames[i] for i in train_idx])
]
alphas = np.logspace(-6, 6, 50)
model.fit(systems_train, spherical_tensormap_train, alphas=alphas)
Evaluate the model¶
We evaluate the model on the test set to check the performance of the model. The API
requires a dictionary of metatensor.torch.ModelOutput objects that specify
the quantity and unit of the output, and whether it is a per_atom quantity or not.
outputs = {
"cookbook::polarizability": ModelOutput(
quantity="polarizability", unit="a.u.", per_atom=False
)
}
systems_test = [
system.to(dtype=torch.float64)
for system in systems_to_torch([ase_frames[i] for i in test_idx])
]
prediction_test = model(systems_test, outputs)
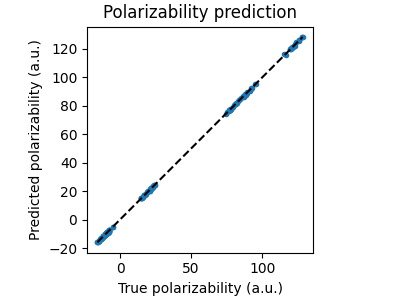
Let us plot the predicted polarizability tensor against the true polarizability tensor for the test set.
true_polarizability = np.concatenate(
[
cartesian_tensormap_test.block(k).values.flatten().numpy()
for k in cartesian_tensormap_test.keys
]
)
predicted_polarizability = np.concatenate(
[
prediction_test["cookbook::polarizability"]
.block(k)
.values.flatten()
.detach()
.numpy()
for k in prediction_test["cookbook::polarizability"].keys
]
)
fig, ax = plt.subplots(1, 1, figsize=(4, 3), constrained_layout=True)
ax.set_aspect("equal")
ax.plot(true_polarizability, predicted_polarizability, ".")
ax.plot(
[true_polarizability.min(), true_polarizability.max()],
[true_polarizability.min(), true_polarizability.max()],
"--k",
)
ax.set_xlabel("True polarizability (a.u.)")
ax.set_ylabel("Predicted polarizability (a.u.)")
ax.set_title("Polarizability prediction")
plt.show()
Export the model as a metatomic model¶
Wrap the model in a
metatomic.torch.AtomisticModel
object.
outputs = {
"cookbook::polarizability": ModelOutput(
quantity="polarizability", unit="a.u.", per_atom=False
)
}
options = ModelEvaluationOptions(
length_unit="angstrom", outputs=outputs, selected_atoms=None
)
model_capabilities = ModelCapabilities(
outputs=outputs,
atomic_types=[1, 6, 7, 16],
interaction_range=4.5,
length_unit="angstrom",
supported_devices=["cpu"],
dtype="float64",
)
atomistic_model = AtomisticModel(model.eval(), ModelMetadata(), model_capabilities)
The model can be saved to disk using the
metatomic.torch.save_atomistic_model() function.
atomistic_model.save("polarizability_model.pt", collect_extensions="extensions")
Use the model as a metatensor calculator¶
The model can be used as a calculator with the
metatomic.torch.MetatomicCalculator class.
atomistic_model = load_atomistic_model(
"polarizability_model.pt", extensions_directory="extensions"
)
mta_calculator = MetatomicCalculator(atomistic_model)
ase_frames = ase.io.read("data/qm7x_reduced_100.xyz", index=":")
ase.Atoms objects do not feature a get_polarizability() method that we
could call to compute the polarizability with our model. The easiest way to compute
the polarizability is then to directly use the MetatomicCalculator.run_model()
method, which takes a list of ase.Atoms objects and a dictionary of
metatomic.torch.ModelOutput objects as input. The output is a
dictionary of metatensor.torch.TensorMap objects.
computed_polarizabilities = []
for frame in ase_frames:
computed_polarizabilities.append(
mta_calculator.run_model(frame, outputs)["cookbook::polarizability"]
.block(0)
.values.squeeze()
.numpy()
)
computed_polarizabilities = np.stack(computed_polarizabilities)
This is essentially the same model as before, wrapped in a stand-alone format
train_mae = np.abs(
(cartesian_polarizability - computed_polarizabilities)[train_idx]
).mean()
test_mae = np.abs(
(cartesian_polarizability - computed_polarizabilities)[test_idx]
).mean()
print(f"""
Model train MAE: {train_mae} a.u.
Model test MAE: {test_mae} a.u.
""")
Model train MAE: 0.049792922120455566 a.u.
Model test MAE: 0.22510172401921538 a.u.
Total running time of the script: (0 minutes 15.744 seconds)