Note
Go to the end to download the full example code.
Fine-tuning the PET-MAD universal potential¶
- Authors:
Davide Tisi @DavideTisi, Zhiyi Wang @0WellyWang0, Cesare Malosso @cesaremalosso, Sofiia Chorna @sofiia-chorna
This example demonstrates how to fine-tune the PET-MAD universal ML potential on a new dataset using metatrain. This allows adapting the model to a specialized task by retraining it on a more focused, domain-specific dataset.
PET-MAD is a universal machine-learning forcefield trained on the MAD dataset that aims to incorporate a very high degree of structural diversity. The model itself is the Point-Edge Transformer (PET), an unconstrained architecture that achieves symmetry compliance through data augmentation during training. You can see an overview of its usage in this introductory example.
The goal of this recipe is to demonstrate the process, not to reach high accuracy. Adjust the dataset size and hyperparameters accordingly if adapting this for an actual application.
# sphinx_gallery_thumbnail_number = 2
import os
import subprocess
from collections import Counter
from glob import glob
import ase.io
import matplotlib.pyplot as plt
import numpy as np
import torch
from huggingface_hub import hf_hub_download
from metatrain.utils.io import model_from_checkpoint
from sklearn.linear_model import LinearRegression
if hasattr(__import__("builtins"), "get_ipython"):
get_ipython().run_line_magic("matplotlib", "inline") # noqa: F821
Preparing for fine-tuning¶
While PET-MAD is trained as a universal model capable of handling a broad range of
atomic environments, fine-tuning it allows to adapt this general-purpose model to a
more specialized task. First, we need to get a checkpoint of the pre-trained PET-MAD
model to start training from it. The checkpoints are stored in the
lab-cosmo/upet Hugging Face repository.
We use huggingface_hub to download it:
checkpoint_cache = hf_hub_download(
repo_id="lab-cosmo/upet",
filename="pet-mad-xs-v1.5.0.ckpt",
subfolder="models",
)
# Copy the checkpoint to the local directory so that it can be referenced
# in the metatrain YAML configuration files.
checkpoint_path = "pet-mad-xs-v1.5.0.ckpt"
if not os.path.exists(checkpoint_path):
import shutil
shutil.copy(checkpoint_cache, checkpoint_path)
Applying an atomic energy correction¶
DFT-calculated energies often contain systematic shifts due to the choice of functional, basis set, or pseudopotentials. If left uncorrected, such shifts can mislead the fine-tuning process.
On this example we use the sampled subset of ethanol structures from rMD17 dataset with PBE/def2-SVP level of theory which is different from PET-MAD v1.5.0, which uses r2SCAN references. We apply a linear correction based on atomic compositions to align our fine-tuning dataset with PET-MAD energy reference. First, we define a helper function to load reference energies from PET-MAD.
def load_reference_energies(checkpoint_path):
"""
Extract atomic reference energies from the PET-MAD checkpoint.
It returns a mapping of elements to their reference energies (eV), e.g.: {'1':
-1.23, '2': -5.67, ...}
"""
checkpoint = torch.load(checkpoint_path, weights_only=False, map_location="cpu")
pet_model = model_from_checkpoint(checkpoint, "finetune")
energy_values = pet_model.additive_models[0].model.weights["energy"].block().values
atomic_numbers = checkpoint["model_data"]["dataset_info"].atomic_types
return dict(zip(atomic_numbers, energy_values))
For demonstration, the dataset is composed only of 100 structures of ethanol. We fit a linear model based on atomic compositions that we use as the energy correction.
dataset = ase.io.read("data/ethanol.xyz", index=":", format="extxyz")
# Extract DFT energies and compositions
dft_energies = [atoms.get_potential_energy() for atoms in dataset]
compositions = [Counter(atoms.get_atomic_numbers()) for atoms in dataset]
elements = sorted({element for composition in compositions for element in composition})
X = np.array([[comp.get(elem, 0) for elem in elements] for comp in compositions])
y = np.array(dft_energies)
# Fit linear model to estimate DFT per-element energy
correction_model = LinearRegression()
correction_model.fit(X, y)
coeffs = dict(zip(elements, correction_model.coef_))
Apply a correction to each structure
def get_compositional_energy(atoms, energy_per_atom):
"""Calculates total energy from atomic composition and per-atom energies"""
counts = Counter(atoms.get_atomic_numbers())
return sum(energy_per_atom[Z] * count for Z, count in counts.items())
# Get reference energies from PET-MAD
ref_energies = load_reference_energies(checkpoint_path)
# Apply correction
for atoms, E_dft in zip(dataset, dft_energies):
E_comp_dft = get_compositional_energy(atoms, coeffs)
E_comp_ref = get_compositional_energy(atoms, ref_energies)
corrected_energy = E_dft - E_comp_dft + E_comp_ref - correction_model.intercept_
atoms.info["energy-corrected"] = corrected_energy.item()
# Split corrected dataset and save it
np.random.seed(42)
indices = np.random.permutation(len(dataset))
n = len(dataset)
n_val = n_test = int(0.1 * n)
n_train = n - n_val - n_test
train = [dataset[i] for i in indices[:n_train]]
val = [dataset[i] for i in indices[n_train : n_train + n_val]]
test = [dataset[i] for i in indices[n_train + n_val :]]
ase.io.write("data/ethanol_train.xyz", train, format="extxyz")
ase.io.write("data/ethanol_val.xyz", val, format="extxyz")
ase.io.write("data/ethanol_test.xyz", test, format="extxyz")
Defines some helper functions¶
We also define a few helper functions to visualize the training results. Each training run generates a log, stored in CSV format in the outputs folder.
def parse_training_log(csv_file):
with open(csv_file, encoding="utf-8") as f:
headers = f.readline().strip().split(",")
cleaned_names = [h.strip().replace(" ", "_") for h in headers]
train_log = np.genfromtxt(
csv_file,
delimiter=",",
skip_header=2,
names=cleaned_names,
)
return train_log
def display_training_curves(train_log, ax=None, style="-", label=""):
"""Plots training and validation losses from the training log"""
if ax is None:
_, (ax1, ax2) = plt.subplots(1, 2, figsize=(8, 4), constrained_layout=True)
else:
ax1, ax2 = ax
ax1.loglog(
train_log["Epoch"],
train_log["training_energy_MAE_per_atom"],
f"r{style}",
label=f"Train. {label}",
)
ax1.loglog(
train_log["Epoch"],
train_log["validation_energy_MAE_per_atom"],
f"b{style}",
label=f"Valid. {label}",
)
ax2.loglog(
train_log["Epoch"],
train_log["training_forces_MAE"],
f"r{style}",
label="Training, F",
)
ax2.loglog(
train_log["Epoch"],
train_log["validation_forces_MAE"],
f"b{style}",
label="Validation, F",
)
ax1.set_xlabel("Epoch")
ax2.set_xlabel("Epoch")
ax1.set_ylabel("Energy MAE (meV)")
ax2.set_ylabel("Force MAE (meV/Å)")
ax1.legend()
return ax1, ax2
Train a model from scratch¶
We first fit a PET model to the corrected dataset to establish a baseline. We use the metatrain utility to train the model. The training is configured in a YAML file, which specifies training, validation and test set, model architecture, optimizer settings, etc. You can learn more about the different settings in the metatrain documentation.
seed: 42
architecture:
name: pet
training:
batch_size: 10
num_epochs: 50
learning_rate: 1e-4
# this needs specifying based on the specific dataset
training_set:
systems:
read_from: "data/ethanol_train.xyz" # path to the finetuning dataset
length_unit: angstrom
targets:
energy:
key: "energy-corrected" # name of the target value
unit: "eV"
validation_set:
systems:
read_from: "data/ethanol_val.xyz"
length_unit: angstrom
targets:
energy:
key: "energy-corrected"
unit: "eV"
test_set:
systems:
read_from: "data/ethanol_test.xyz"
length_unit: angstrom
targets:
energy:
key: "energy-corrected"
unit: "eV"
To launch training, you just need to run the following command in the terminal:
mtt train <options.yaml> [-o <output.pt>]
Or from Python:
subprocess.run(
["mtt", "train", "from_scratch_options.yaml", "-o", "from_scratch-model.pt"],
check=True,
)
CompletedProcess(args=['mtt', 'train', 'from_scratch_options.yaml', '-o', 'from_scratch-model.pt'], returncode=0)
The training logs are stored in the outputs/ directory, with a subdirectory
named by the date and time of the training run. The model checkpoint is saved as
model.ckpt and the exported model as model.pt, unless specified otherwise
with the -o option.
We can load the latest training log and visualize the training curves - we will use them later to compare the fine-tuning results. It is clear that training is not converged, and the learning rate is not optimal – you can try to adjust the parameters and run for longer.
csv_file = sorted(glob("outputs/*/*/train.csv"))[-1]
from_scratch_log = parse_training_log(csv_file)
display_training_curves(from_scratch_log)
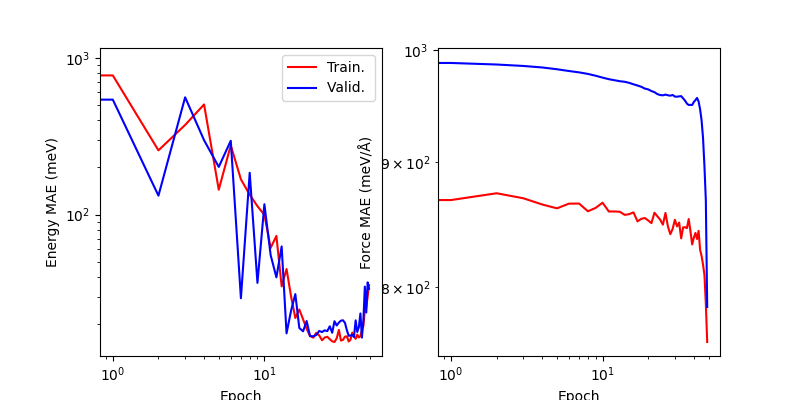
(<Axes: xlabel='Epoch', ylabel='Energy MAE (meV)'>, <Axes: xlabel='Epoch', ylabel='Force MAE (meV/Å)'>)
Simple model fine-tuning¶
Having prepared the dataset and fitted a baseline model “from scratch”,
we proceed with the training of a fine-tuned model. To this end, we also use the
metatrain package. There are multiple strategies to apply
fine-tuning, each described in the documentation.
In this example we demonstrate a basic full fine-tuning strategy, which adapts all
model weights to the new dataset starting from the pre-trained PET-MAD checkpoint. The
process is configured by setting appropriate settings in the YAML options file.
seed: 42
architecture:
name: "pet"
training:
batch_size: 10
num_epochs: 50 # very short period for demonstration
learning_rate: 1e-5 # small learning rate to stabilize training
finetune:
method: "full" # use fine-tuning strategy
read_from: pet-mad-xs-v1.5.0.ckpt # path to the pretrained checkpoint to start from
training_set:
systems:
read_from: "data/ethanol_train.xyz" # path to the finetuning dataset
length_unit: angstrom
targets:
energy:
key: "energy-corrected" # name of the target value
unit: "eV"
validation_set:
systems:
read_from: "data/ethanol_val.xyz"
length_unit: angstrom
targets:
energy:
key: "energy-corrected"
unit: "eV"
test_set:
systems:
read_from: "data/ethanol_test.xyz"
length_unit: angstrom
targets:
energy:
key: "energy-corrected"
unit: "eV"
subprocess.run(
["mtt", "train", "full_ft_options.yaml", "-o", "fine_tune-model.pt"], check=True
)
CompletedProcess(args=['mtt', 'train', 'full_ft_options.yaml', '-o', 'fine_tune-model.pt'], returncode=0)
Comparing the model tranined from scratch (dashed lines) and the fine-tuned one (full lines), it is clear that fine-tuning from PET-MAD weights leads to much better zero-shot accuracy, and more consistent learning dynamics. Obviously it may be possible to tweak differently, and it is not unlikely that a large single-purpose training set and long training time might lead to better validation error than performing fine tuning.
csv_file = sorted(glob("outputs/*/*/train.csv"))[-1]
fine_tune_log = parse_training_log(csv_file)
ax = display_training_curves(fine_tune_log, label="Fine tuning")
display_training_curves(from_scratch_log, ax=ax, style="--", label="From scratch")
ax[0].set_ylim(1, 1000)
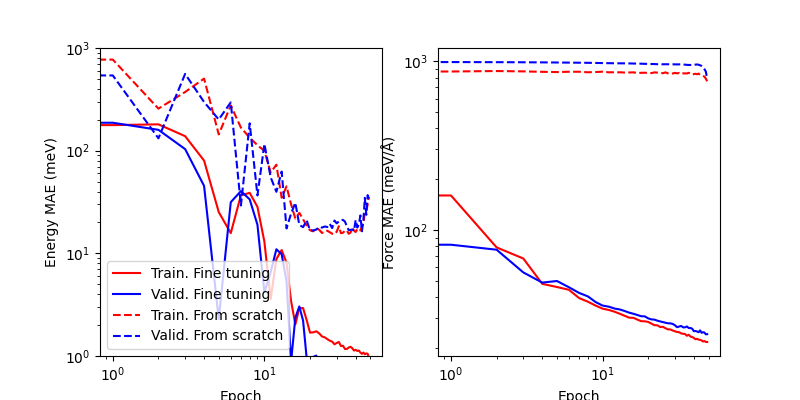
(1, 1000)
Model evaluation¶
After the training, mtt train outputs the fine_tune-model.ckpt
and fine_tune-model.pt (exported fine-tuned model) files in both the
current directory and in output/YYYY-MM-DD/HH-MM-SS/.
These can be used together with metatrain to evaluate the model on a (potentially different) dataset. The evaluation is configured in a YAML file, which specifies the dataset to use, and the metrics to compute.
systems:
read_from: data/ethanol_test.xyz
targets:
energy:
key: energy-corrected
unit: eV
The evaluation can be run from the command line:
mtt eval fine_tune-model.pt model_eval.yaml
Or from Python:
result = subprocess.run(
["mtt", "eval", "fine_tune-model.pt", "model_eval.yaml"],
check=True,
capture_output=True,
text=True,
)
print(result.stdout)
[2026-03-04 06:55:25][INFO] - Logging to file is disabled.
[2026-03-04 06:55:25][INFO] - Package version: 2026.2.1
[2026-03-04 06:55:25][INFO] - Package directory: /home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-finetuning/lib/python3.12/site-packages/metatrain
[2026-03-04 06:55:25][INFO] - Working directory: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/pet-finetuning
[2026-03-04 06:55:25][INFO] - Executed command: mtt eval fine_tune-model.pt model_eval.yaml
[2026-03-04 06:55:25][INFO] - Setting up evaluation set.
[2026-03-04 06:55:25][INFO] - Evaluating dataset
[2026-03-04 06:55:25][INFO] - Forces found in section 'energy-corrected', we will use this gradient to train the model
[2026-03-04 06:55:25][WARNING] - No stress found in section 'energy-corrected'.
[2026-03-04 06:55:25][INFO] - Running on device cpu with dtype torch.float32
[2026-03-04 06:55:27][INFO] - energy RMSE (per atom): 2.2220 meV | energy MAE (per atom): 1.9075 meV | forces RMSE: 40.887 meV/A | forces MAE: 31.021 meV/A
[2026-03-04 06:55:27][INFO] - Evaluation time: 0.16 s [1.7581 ± 0.0599 ms per atom]
Total running time of the script: (2 minutes 40.055 seconds)