Note
Go to the end to download the full example code.
Uncertainty Quantification with PET-MAD¶
- Authors:
Johannes Spies @johannes-spies
This recipe demonstrates three ways of computing errors on the outputs of ML potential-driven simulations, using as an example the PET-MAD model and its built-in uncertainty quantification (UQ) capabilities.
In particular, it demonstrates:
Estimating uncertainties for single-point calculations on a full validation dataset.
Computing energies in simple functions of energy predictions, namely the value of vacancy formation energies
Propagating errors from energy predictions to thermodynamic averages computed over a constant-temperature MD simulation.
For more information on PET-MAD, have a look at Mazitov et al., 2025. The LLPR uncertainties are introduced in Bigi et al., 2024. For more information on dataset calibration and error propagation, see Imabalzano et al., 2021.
Optional: Adding UQ to a Model¶
Models compatible with metatomic can be equipped with UQ capabilities through the LLPRUncertaintyModel wrapper included with metatrain. For running this recipe, you can use a prebuilt model (the example itself downloads a model from Hugging Face). For adding UQ support to an existing model, have a look at the following scaffold. For more information on loading a dataset with the infrastructure, have a look at this section of the documentation. The pseudocode below also shows how to create an ensemble model from the last-layer parameters of a model.
from metatrain.utils.llpr import LLPRUncertaintyModel
from metatomic.torch import AtomisticModel, ModelMetadata
# You need to provide a model and datasets (wrapped in PyTorch dataloaders).
model = ...
dataloaders = {"train": ..., "val": ...}
# Wrap the model in a module capable of estimating uncertainties, estimate the
# inverse covariance on the training set, and calibrate the model on the validation
# set.
llpr_model = LLPRUncertaintyModel(model)
llpr_model.compute_covariance(dataloaders["train"])
llpr_model.compute_inverse_covariance(regularizer=1e-4)
llpr_model.calibrate(dataloaders["val"])
# In the next step, we show how to enable ensembles in PET-MAD. For that, it is
# necessary to extract the last-layer parameters of the model. The ensemble
# generation expects the parameters in a flat-vector format.
last_layer_parameters = ...
# Generate an ensemble with 128 members to compare ensemble uncertainties to LLPR
# scores.
llpr_model.generate_ensemble({"energy": last_layer_parameters}, 128)
# Save the model to disk using metatomic.
exported_model = AtomisticModel(
llpr_model.eval(),
ModelMetadata(),
llpr_model.capabilities,
)
exported_model.save("models/model-with-llpr.pt")
Getting Started¶
At the bottom of the page, you’ll find a ZIP file containing the whole example. Note that it comes with an environment.yml file specifying all dependencies required to execute the script.
import os
import subprocess
from urllib.request import urlretrieve
import ase.geometry.rdf
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
from ase import Atoms
from ase.filters import FrechetCellFilter
from ase.io.cif import read_cif
from ase.optimize.bfgs import BFGS
from ipi.utils.scripting import InteractiveSimulation
from metatomic.torch import ModelOutput
from metatomic.torch.ase_calculator import MetatomicCalculator
from metatrain.utils.data import Dataset, read_systems, read_targets
from metatrain.utils.data.system_to_ase import system_to_ase
Model Loading¶
All examples require a PET-MAD model with ensemble and LLPR prediction. The following code loads a pre-trained model using the ASE-compatible calculator wrapper. Using the calculator instead of calling the model directly conveniently hides computing neighbor lists in the calculator.
if not os.path.exists("models/pet-mad-latest-llpr.pt"):
os.makedirs("models", exist_ok=True)
urlretrieve(
"https://huggingface.co/jospies/pet-mad-llpr/resolve/main/"
"pet-mad-latest-llpr.pt?download=true",
"models/pet-mad-latest-llpr.pt",
)
calculator = MetatomicCalculator("models/pet-mad-latest-llpr.pt", device="cpu")
Uncertainties on a Dataset¶
This first example shows how to use PET-MAD to estimate uncertainties on a reference dataset. We use a reduced version (because of limited compute power in the CI runner) of the MAD validation set.
For this, we first download the correspond MAD validation dataset record from Materials Cloud. Then, we prepare the dataset and pass it through the model. In the final step, we visualize the predicted uncertainties and compare them to a ground truth method.
if not os.path.exists("data/mad-val-100.xyz"):
os.makedirs("data", exist_ok=True)
urlretrieve(
"https://huggingface.co/jospies/pet-mad-llpr/resolve/main/mad-val-100.xyz"
"?download=true",
"data/mad-val-100.xyz",
)
# Read the dataset's structures.
systems = read_systems("data/mad-val-100.xyz")
# Read the dataset's targets.
target_config = {
"energy": {
"quantity": "energy",
"read_from": "data/mad-val-100.xyz",
"reader": "ase",
"key": "energy",
"unit": "kcal/mol",
"type": "scalar",
"per_atom": False,
"num_subtargets": 1,
"forces": False,
"stress": False,
"virial": False,
},
}
targets, infos = read_targets(target_config) # type: ignore
# Wrap in a `metatrain` compatible way.
dataset = Dataset.from_dict({"system": systems, **targets})
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/pet-mad-uq/lib/python3.12/site-packages/metatrain/utils/data/readers/ase.py:62: UserWarning: A conversion to `System` was requested for an `ase.Atoms` object with one or more non-zero cell vectors but where the corresponding boundary conditions are set to `False`. The corresponding cell vectors will be set to zero.
systems = systems_to_torch(ase_atoms, dtype=torch.float64)
After preparation, the dataset can be passed through the model using the calculator to obtain energy predictions and LLPR scores.
# Convert the systems to an ASE-native `Atoms` object
systems = [system_to_ase(sample["system"]) for sample in dataset]
outputs = {
# Request the uncertainty in the atomic energy predictions
"energy": ModelOutput(), # (Needed to request the uncertainties)
"energy_uncertainty": ModelOutput(),
}
results = calculator.run_model(systems, outputs)
# Extract the requested results
predicted_energies = results["energy"][0].values.squeeze()
predicted_uncertainties = results["energy_uncertainty"][0].values.squeeze()
Compute the true prediction error by comparing the predicted energy to the reference value from dataset.
# Reference values from dataset.
ground_truth_energies = torch.stack(
[sample["energy"][0].values.squeeze() for sample in dataset]
)
# Compute squared distance between predicted energy and reference value.
empirical_errors = torch.abs(predicted_energies - ground_truth_energies)
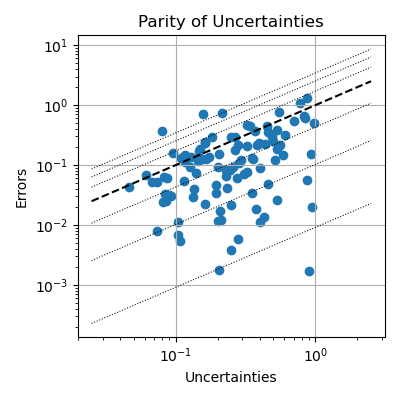
After gathering predicted uncertainties and computing ground truth error metrics, we can compare them to each other. Similar to figure S4 of the PET-MAD paper, we present the data in using a parity plot. For more information about interpreting this type of plot, see Appendix F.7 of Bigi et al., 2024. Note that both the x- and the y-axis use a logarithmic scale, which is more suitable for inspecting uncertainty values. Because we are using a heavily reduced dataset (only 100 structures) from the MAD validation set, the parity plot looks very sparse.
# Hard-code the zoomed in region of the plot and iso-lines.
quantile_lines = [0.00916, 0.10256, 0.4309805, 1.71796, 2.5348, 3.44388]
min_val, max_val = 2.5e-2, 2.5
# Create the parity plot.
plt.figure(figsize=(4, 4))
plt.grid()
plt.gca().set(
title="Parity of Uncertainties",
ylabel="Errors",
xlabel="Uncertainties",
)
plt.loglog()
# Plot iso lines.
plt.plot([min_val, max_val], [min_val, max_val], ls="--", c="k")
for factor in quantile_lines:
plt.plot([min_val, max_val], [factor * min_val, factor * max_val], "k:", lw=0.75)
# Add actual samples.
plt.scatter(predicted_uncertainties, empirical_errors)
plt.tight_layout()
Uncertainties in Vacancy Formation Energies¶
One can use ensemble uncertainty quantification to estimate the error in predicting vacancy formation energies, which we show in this example.
In this part, we use an aluminum crystal as an example system. The structure file can be downloaded from Material Project as a .cif file. We’ve included such a file with the recipe.
The following code loads the structure, computes the energy before creating a defect, creates a defect, runs a structural optimization, and computes the energy after the optimization. The energy difference can be used to estimate the vacancy formation energy.
# Load the crystal from the Materials Project and create a supercell (not strictly
# necessary).
crystal_structure = "data/Al_mp-134_conventional_standard.cif"
atoms: Atoms = read_cif(crystal_structure) # type: ignore
supercell = atoms * 2
supercell.calc = calculator
N = len(supercell) # store the number of atoms
We now compute the vacancy formation energy by keeping track of the ensemble energies at different stages. Note that calling .get_potential_energy() on an Atoms object triggers computing the ensemble values.
# Get ensemble energy before creating the vacancy
outputs = ["energy", "energy_uncertainty", "energy_ensemble"]
outputs = {o: ModelOutput() for o in outputs}
results = calculator.run_model(supercell, outputs)
bulk = results["energy_ensemble"][0].values
# Remove an atom (last atom in this case) to create a vacancy
i = -1
supercell.pop(i)
# Get ensemble energy right after creating the vacancy
results = calculator.run_model(supercell, outputs)
right_after_vacancy = results["energy_ensemble"][0].values
# Run structural optimization optimizing both positions and cell layout.
ecf = FrechetCellFilter(supercell)
bfgs = BFGS(ecf) # type: ignore
bfgs.run()
# get ensembele energy after optimization
results = calculator.run_model(supercell, outputs)
vacancy = results["energy_ensemble"][0].values
Step Time Energy fmax
BFGS: 0 14:52:31 -122.585747 0.130144
BFGS: 1 14:52:31 -122.589012 0.122949
BFGS: 2 14:52:31 -122.620888 0.096033
BFGS: 3 14:52:31 -122.621590 0.095035
BFGS: 4 14:52:31 -122.629784 0.078909
BFGS: 5 14:52:32 -122.633263 0.069313
BFGS: 6 14:52:32 -122.635338 0.062530
BFGS: 7 14:52:32 -122.636513 0.060592
BFGS: 8 14:52:32 -122.639427 0.065045
BFGS: 9 14:52:32 -122.644012 0.058950
BFGS: 10 14:52:32 -122.649384 0.035773
Compute vacancy formation energy for each ensemble member.
vacancy_formation = vacancy - (N - 1) / N * bulk
Put all ensemble energies in a dataframe and compute the desired statistics.
# This dataframe contains each stage's energies in a single column.
named_stages = [
("Before creating vacancy", bulk),
("Right after creating vacancy", right_after_vacancy),
("Energy of optimized vacancy", vacancy),
("Vacancy formation energy", vacancy_formation),
]
df = pd.DataFrame.from_dict(
{
# Convert the PyTorch tensors to flat NumPy vectors
k: v.detach().numpy().squeeze()
for k, v in named_stages
}
)
# Compute statistics (mean, variance, and standard deviation) on the ensemble energies.
df = pd.DataFrame(dict(mean=df.mean(), var=df.var(), std=df.std()))
df
Uncertainty Propagation with MD¶
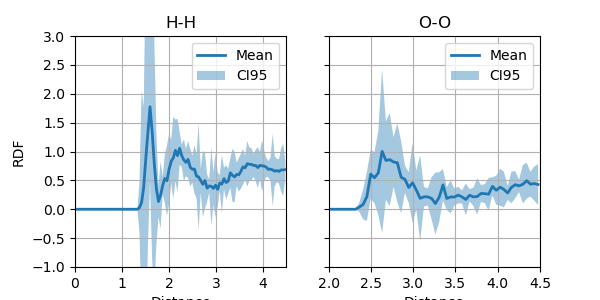
This example shows how to use i-PI to propagate error estimates from an ensemble to output observables. In this example, we use a box with period boundary conditions housing 32 water molecules. As an observable, we inspect the Radial Distribution Function (RDF) between hydrogen-hydrogen and oxygen-oxygen bonds.
First, we run a simulation with i-PI generating a trajectory and logging other metrics. The trajectory and committee energies can be used in a subsequent postprocessing step to obtain RDFs using ASE. These can be re-weighted to propagate errors from the committee uncertainties to the observed RDFs.
Note also that we set a uncertainty_threshold option in the driver. When running from the command line, this will output a warning every time one of the atomic energy is estimated to have an uncertainty above that threshold (in eV/atom).
# Load configuration and run simulation.
with open("data/h2o-32.xml") as f:
xml_input = f.read()
# prints the relevant sections of the input file
print(xml_input[:883][-334:])
sim = InteractiveSimulation(xml_input)
rue">
<pes>metatomic</pes>
<parameters>
{ template:data/h2o-32.xyz,model:models/pet-mad-latest-llpr.pt,device:cpu,
non_conservative:False,energy_ensemble:True,
check_consistency:False,
uncertainty_threshold:7.0e-2
}
</parameters>
</ffdirect>
<system>
<initialize nbead
@system: Initializing system object
@simulation: Initializing simulation object
@ RANDOM SEED: The seed used in this calculation was 23658
@init_file: Initializing from file data/h2o-32.xyz. Dimension: length, units: angstrom, cell_units: automatic
@init_file: Initializing from file data/h2o-32.xyz. Dimension: length, units: automatic, cell_units: automatic
@init_file: Initializing from file data/h2o-32.xyz. Dimension: length, units: automatic, cell_units: automatic
@init_file: Initializing from file data/h2o-32.xyz. Dimension: length, units: automatic, cell_units: automatic
@init_file: Initializing from file data/h2o-32.xyz. Dimension: length, units: automatic, cell_units: automatic
@initializer: Resampling velocities at temperature 300.0 kelvin
--- begin input file content ---
<simulation verbosity='medium'>
<output prefix='h2o-32'>
<properties filename='out' stride='10'>[ time{picosecond}, temperature{kelvin}, kinetic_md ]</properties>
<trajectory filename='pos' format='ase' stride='10'>positions</trajectory>
<trajectory filename='committee_pot' stride='10' extra_type='committee_pot'>extras</trajectory>
<trajectory filename='atomic_error' stride='10' extra_type='energy_uncertainty'>extras</trajectory>
</output>
<prng>
<seed>23658</seed>
</prng>
<ffdirect name='nocons' pbc='true'>
<pes>metatomic</pes>
<parameters>{ template:data/h2o-32.xyz,model:models/pet-mad-latest-llpr.pt,device:cpu,
non_conservative:False,energy_ensemble:True,
check_consistency:False,
uncertainty_threshold:7.0e-2
}</parameters>
</ffdirect>
<system>
<initialize nbeads='1'>
<file mode='xyz' units='angstrom'>data/h2o-32.xyz</file>
<velocities mode='thermal' units='kelvin'>300</velocities>
</initialize>
<forces>
<force forcefield='nocons'>
</force>
</forces>
<ensemble>
<temperature units='kelvin'>300</temperature>
</ensemble>
<motion mode='dynamics'>
<fixcom>True</fixcom>
<dynamics mode='nvt'>
<timestep units='femtosecond'>0.5</timestep>
<thermostat mode='langevin'>
<tau units='femtosecond'>50</tau>
</thermostat>
</dynamics>
</motion>
</system>
</simulation>
--- end input file content ---
@system.bind: Binding the forces
Run the simulation.
# NB: To get better estimates, set this to a higher number (perhaps 10000) to
# run the simulation for a longer time.
sim.run(400)
@simulation.run: Average timings at MD step 0. t/step: 1.22158e+00
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0700 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0703 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0705 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0705 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0703 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0700 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0700 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
@simulation.run: Average timings at MD step 100. t/step: 4.85674e-01
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0705 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0703 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0703 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0700 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0702 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0703 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0705 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0717 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0717 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0717 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
@simulation.run: Average timings at MD step 200. t/step: 4.86593e-01
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0718 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0719 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0721 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0722 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0723 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0725 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0727 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0729 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0730 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0731 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0732 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0732 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0731 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0730 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0729 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0727 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0726 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0725 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0725 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0724 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0723 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0723 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0722 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0722 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0721 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0721 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0721 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0721 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0720 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0720 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0720 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0719 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0719 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0717 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
@simulation.run: Average timings at MD step 300. t/step: 4.99329e-01
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0708 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0705 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0707 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0716 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0715 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0710 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0713 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0714 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0712 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0711 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0709 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0706 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0704 eV/atom exceeds the selected threshold of 0.0700 eV/atom
!W! the estimated atomic energy uncertainty 0.0701 eV/atom exceeds the selected threshold of 0.0700 eV/atom
Load the trajectories and compute the per-frame RDFs Note that ASE applies a weird normalization to the partial RDFs, which require a correction to recover the usual asymptotic behavior at large distances.
frames: list[Atoms] = ase.io.read("h2o-32.pos_0.extxyz", ":") # type: ignore
# Our simulation should only include water molecules. (types: hydrogen=1, oxygen=8)
assert set(frames[0].numbers.tolist()) == set([1, 8])
# Compute the RDF of each frame (for H-H and for O-O)
num_bins = 250
rdfs_hh = []
rdfs_oo = []
xs = None
for atoms in frames:
# Compute H-H distances
bins, xs = ase.geometry.rdf.get_rdf( # type: ignore
atoms, 4.5, num_bins, elements=[1, 1]
)
# smoothen the RDF a bit (not enough data...)
bins[2:-2] = (
bins[:-4] * 0.1
+ bins[1:-3] * 0.2
+ bins[2:-2] * 0.4
+ bins[3:-1] * 0.2
+ bins[4:] * 0.1
)
rdfs_hh.append(bins * 3.0 / 2.0) # correct ASE normalization
# Compute O-O distances
bins, xs = ase.geometry.rdf.get_rdf( # type: ignore
atoms, 4.5, num_bins, elements=[8, 8]
)
bins[2:-2] = (
bins[:-4] * 0.1
+ bins[1:-3] * 0.2
+ bins[2:-2] * 0.4
+ bins[3:-1] * 0.2
+ bins[4:] * 0.1
)
rdfs_oo.append(bins * 3.0) # correct ASE normalization
rdfs_hh = np.stack(rdfs_hh, axis=0)
rdfs_oo = np.stack(rdfs_oo, axis=0)
Run the i-PI re-weighting utility as a post-processing step.
# Save RDFs such that they can be read from i-PI.
np.savetxt("h2o-32_rdfs_h-h.txt", rdfs_hh)
np.savetxt("h2o-32_rdfs_o-o.txt", rdfs_oo)
# Run the re-weighting tool from i-PI for H-H and O-O
for ty in ["h-h", "o-o"]:
infile = f"h2o-32_rdfs_{ty}.txt"
outfile = f"h2o-32_rdfs_{ty}_reweighted.txt"
cmd = (
f"i-pi-committee-reweight h2o-32.committee_pot_0 {infile} --input"
" data/h2o-32.xml"
)
print("Executing command:", "\t" + cmd, sep="\n")
cmd = cmd.split()
with open(outfile, "w") as out:
process = subprocess.run(cmd, stdout=out)
Executing command:
i-pi-committee-reweight h2o-32.committee_pot_0 h2o-32_rdfs_h-h.txt --input data/h2o-32.xml
Executing command:
i-pi-committee-reweight h2o-32.committee_pot_0 h2o-32_rdfs_o-o.txt --input data/h2o-32.xml
Load and display the RDFs after re-weighting. Note that the results might not noisy due to the small number of MD steps.
# Load the reweighted RDFs.
rdfs_hh_reweighted = np.loadtxt("h2o-32_rdfs_h-h_reweighted.txt")
rdfs_oo_reweighted = np.loadtxt("h2o-32_rdfs_o-o_reweighted.txt")
# Extract columns.
rdfs_hh_reweighted_mu = rdfs_hh_reweighted[:, 0]
rdfs_hh_reweighted_err = rdfs_hh_reweighted[:, 1]
rdfs_hh_reweighted_committees = rdfs_hh_reweighted[:, 2:]
rdfs_oo_reweighted_mu = rdfs_oo_reweighted[:, 0]
rdfs_oo_reweighted_err = rdfs_oo_reweighted[:, 1]
rdfs_oo_reweighted_committees = rdfs_oo_reweighted[:, 2:]
# Display results.
fig, axs = plt.subplots(figsize=(6, 3), sharey=True, ncols=2)
for title, ax, mus, std, xlim in [
("H-H", axs[0], rdfs_hh_reweighted_mu, rdfs_hh_reweighted_err, (1.0, 4.5)),
("O-O", axs[1], rdfs_oo_reweighted_mu, rdfs_oo_reweighted_err, (2.0, 4.5)),
]:
ylabel = "RDF" if title == "H-H" else None
ax.set(title=title, xlabel="Distance", ylabel=ylabel, xlim=xlim, ylim=(-0.1, 3.7))
ax.grid()
ax.plot(xs, mus, label="Mean", lw=0.5)
z95 = 1.96
rdfs_ci95 = (mus - z95 * std, mus + z95 * std)
ax.fill_between(xs, *rdfs_ci95, alpha=0.5, label="CI95")
ax.legend()
Total running time of the script: (3 minutes 35.206 seconds)