Universal ML models¶
This section contains recipes based on “universal” ML models - i.e. models that are trained to make prediction across a a substantial fraction of chemical and structural space.
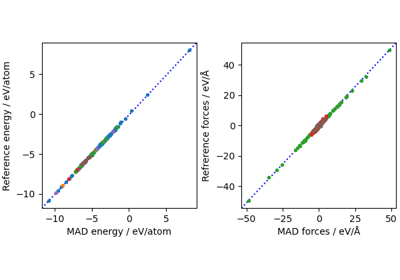
The PET-MAD universal potential
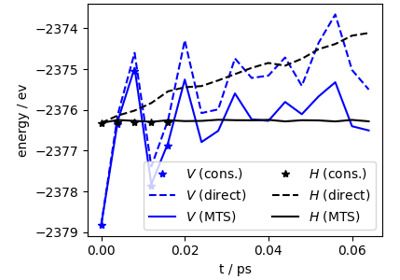
MD using direct-force predictions with PET-MAD
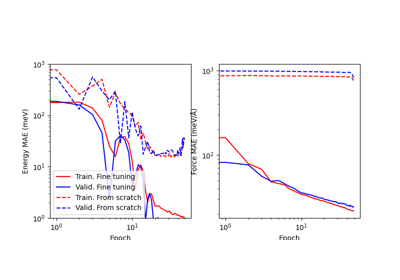
Fine-tuning the PET-MAD universal potential
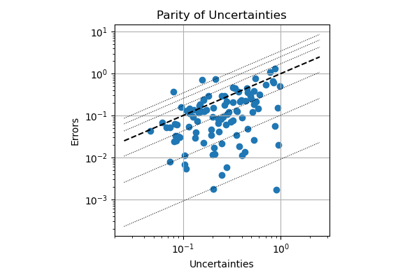
Uncertainty Quantification with PET-MAD
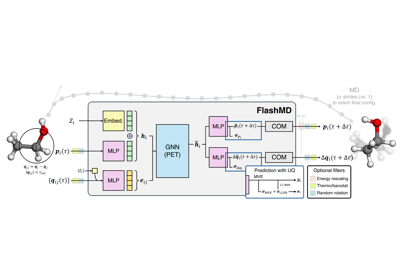
Long-stride trajectories with a universal FlashMD model
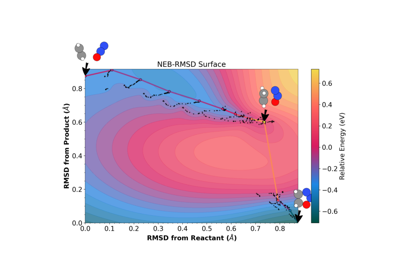
Finding Reaction Paths with eOn and a Metatomic Potential