Note
Go to the end to download the full example code.
Finding Reaction Paths with eOn and a Metatomic Potential¶
- Authors:
Rohit Goswami @HaoZeke, Hanna Tuerk @HannaTuerk, Arslan Mazitov @abmazitov, Michele Ceriotti @ceriottim
This example describes how to find the reaction pathway for oxadiazole formation from N₂O and ethylene. We will use the PET-MAD metatomic model to calculate the potential energy and forces.
The primary goal is to contrast a standard Nudged Elastic Band (NEB) calculation using the atomic simulation environment (ASE) with more sophisticated methods available in the eOn package. For even a relatively simple reaction like this, a basic NEB implementation can struggle to converge or may time out. We will show how eOn’s advanced features, such as energy-weighted springs and mixing in single-ended dimer search steps, can efficiently locate and refine the transition state along the path.
Our approach will be:
Set up the PET-MAD metatomic calculator.
Use ASE to generate an initial IDPP reaction path.
Illustrate the limitations of a standard NEB calculation in ASE.
Refine the path and locate the transition state saddle point using eOn’s optimizers, including energy-weighted springs and the dimer method.
Visualize the final converged pathway.
Demonstrate endpoint relaxation with eOn
Importing Required Packages¶
First, we import all the necessary python packages for this task.
By convention, all import
statements are at the top of the file.
import contextlib
import os
import subprocess
import sys
from pathlib import Path
import ase
import ase.io as aseio
import ira_mod
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
from ase.mep import NEB
from ase.optimize import LBFGS
from ase.visualize import view
from ase.visualize.plot import plot_atoms
from metatomic.torch.ase_calculator import MetatomicCalculator
from rgpycrumbs.eon.helpers import write_eon_config
from rgpycrumbs.run.jupyter import run_command_or_exit
# sphinx_gallery_thumbnail_number = 4
Obtaining the Foundation Model - PET-MAD¶
PET-MAD is an instance of a point edge transformer model trained on
the diverse MAD dataset
which learns equivariance through data driven measures
instead of having equivariance baked in [1]. In turn, this enables
the PET model to have greater design space to learn over. Integration in
Python and the C++ eOn client occurs through the metatomic software [2],
which in turn relies on the atomistic machine learning toolkit build
over metatensor. Essentially using any of the metatomic models involves
grabbing weights off of HuggingFace and loading them with
metatomic before running the
engine of choice.
repo_id = "lab-cosmo/upet"
tag = "v1.1.0"
url_path = f"models/pet-mad-s-{tag}.ckpt"
fname = Path(url_path.replace(".ckpt", ".pt"))
url = f"https://huggingface.co/{repo_id}/resolve/main/{url_path}"
fname.parent.mkdir(parents=True, exist_ok=True)
subprocess.run(
[
"mtt",
"export",
url,
"-o",
fname,
],
check=True,
)
print(f"Successfully exported {fname}.")
Successfully exported models/pet-mad-s-v1.1.0.pt.
Nudged Elastic Band (NEB)¶
Given two known configurations on a potential energy surface (PES), often one wishes to determine the path of highest probability between the two. Under the harmonic approximation to transition state theory, connecting the configurations (each point representing a full molecular structure) by a discrete set of images allows one to evolve the path under an optimization algorithm, and allows approximating the reaction to three states: the reactant, product, and transition state.
The location of this transition state (≈ the point with the highest energy along this path) determines the barrier height of the reaction. This saddle point can be found by transforming the second derivatives (Hessian) to step along the softest mode. However, an approximation which is free from explicitly finding this mode involves moving the highest image of a NEB path: the “climbing” image.
Mathematically, the saddle point has zero first derivatives and a single negative eigenvalue. The climbing image technique moves the highest energy image along the reversed NEB tangent force, avoiding the cost of full Hessian diagonalization used in single-ended methods [3].
Concretely, in this example, we will consider a reactant and product state, for oxadiazole formation, namely N₂O and ethylene.
reactant = aseio.read("data/min_reactant.con")
product = aseio.read("data/min_product.con")
We can visualize these structures using ASE.
fig, (ax1, ax2) = plt.subplots(1, 2)
plot_atoms(reactant, ax1, rotation=("-90x,0y,0z"))
plot_atoms(product, ax2, rotation=("-90x,0y,0z"))
ax1.text(0.3, -1, "reactant")
ax2.text(0.3, -1, "product")
ax1.set_axis_off()
ax2.set_axis_off()
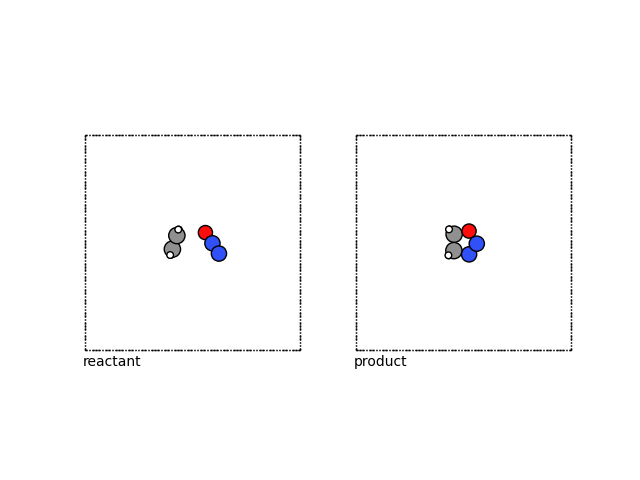
Endpoint minimization¶
For finding reaction pathways, the endpoints should be minimized. We provide initial configurations which are already minimized, but in order to see how to relax endpoints with eOn, please have a look at the end of this tutorial.
Initial path generation¶
To begin an NEB method, an initial path is required, the optimal construction of which still forms an active area of research. The simplest approximation to an initial path for NEB methods linearly interpolate between the two known configurations building on intuition developed from “drag coordinate” methods. This may break bonds or otherwise also unphysically pass atoms through each other, similar to the effect of incorrect permutations. To ameliorate this effect, the NEB algorithm is often started from the linear interpolation but then the path is optimized on a surrogate potential energy surface, commonly something cheap and analytic, like the IDPP (Image dependent pair potential, [5]) which provides a surface based on bond distances, and thus preventing atom-in-atom collisions.
Here we use the IDPP from ASE to setup the initial path. You can find more information about this method in the corresponding ASE tutorial or in the original IDPP publication by S. Smidstru et al. . A brief pedagogical discussion of the transition state methods may be found on the Rowan blog, though the software is proprietary there.
N_INTERMEDIATE_IMGS = 10
# total includes the endpoints
TOTAL_IMGS = N_INTERMEDIATE_IMGS + 2
images = [reactant]
images += [reactant.copy() for img in range(N_INTERMEDIATE_IMGS)]
images += [product]
neb = NEB(images)
neb.interpolate("idpp")
We don’t cover subtleties in setting the number of images, typically too many intermediate images may cause kinks but too few will be unable to resolve the tangent to any reasonable quality.
For eOn, we write the initial path to a file called idppPath.dat.
output_dir = "path"
os.makedirs(output_dir, exist_ok=True)
output_files = [f"{output_dir}/{num:02d}.con" for num in range(TOTAL_IMGS)]
for outfile, img in zip(output_files, images):
ase.io.write(outfile, img)
print(f"Wrote {len(output_files)} IDPP images to '{output_dir}/'.")
summary_file_path = "idppPath.dat"
with open(summary_file_path, "w") as f:
for filepath in output_files:
abs_path = os.path.abspath(filepath)
f.write(f"{abs_path}\n")
print(f"Wrote absolute paths to '{summary_file_path}'.")
Wrote 12 IDPP images to 'path/'.
Wrote absolute paths to 'idppPath.dat'.
Running NEBs¶
We will now consider actually running the Nudged Elastic Band with different codes.
ASE and Metatomic¶
We first consider using metatomic with the ASE calculator.
# define the calculator
def mk_mta_calc():
return MetatomicCalculator(
fname,
device="cpu",
non_conservative=False,
uncertainty_threshold=0.001,
)
# set calculators for images
ipath = [reactant] + [reactant.copy() for img in range(10)] + [product]
for img in ipath:
img.calc = mk_mta_calc()
print(img.calc._model.capabilities().outputs)
neb = NEB(ipath, climb=True, k=5, method="improvedtangent")
neb.interpolate("idpp")
# store initial path guess for plotting
initial_energies = [img.get_potential_energy() for img in ipath]
# setup the NEB clalculation
optimizer = LBFGS(neb, trajectory="A2B.traj", logfile="opt.log")
conv = optimizer.run(fmax=0.01, steps=100)
print("Check if calculation has converged:", conv)
if conv:
print(neb)
final_energies = [i.get_potential_energy() for i in ipath]
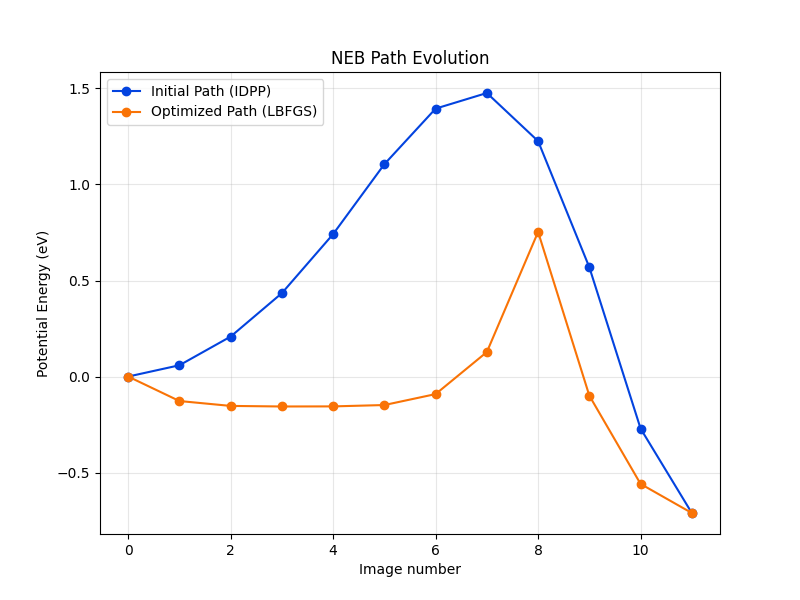
# Plot initial and final path
plt.figure(figsize=(8, 6))
# Initial Path (Blue)
plt.plot(
initial_energies - initial_energies[0],
"o-",
label="Initial Path (IDPP)",
color="xkcd:blue",
)
# Final Path (Orange)
plt.plot(
final_energies - initial_energies[0],
"o-",
label="Optimized Path (LBFGS)",
color="xkcd:orange",
)
# Metadata
plt.xlabel("Image number")
plt.ylabel("Potential Energy (eV)")
plt.legend()
plt.grid(True, alpha=0.3)
plt.title("NEB Path Evolution")
plt.show()
{'energy': <torch.ScriptObject object at 0x56303ff192f0>, 'features': <torch.ScriptObject object at 0x56303c9482b0>, 'mtt::aux::energy_last_layer_features': <torch.ScriptObject object at 0x56303fc0f320>, 'mtt::aux::non_conservative_forces_last_layer_features': <torch.ScriptObject object at 0x563040560fe0>, 'mtt::aux::non_conservative_stress_last_layer_features': <torch.ScriptObject object at 0x5630404ef6e0>, 'non_conservative_forces': <torch.ScriptObject object at 0x56303e0a9ee0>, 'non_conservative_stress': <torch.ScriptObject object at 0x56303f15dbe0>}
Check if calculation has converged: True
<ase.mep.neb.NEB object at 0x7f8cb9ce6e90>
In the 100 NEB steps we took, the structure did unfortunately not converge. The metatomic calculator for PET-MAD v1.0.2 provides LLPR based energy uncertainties. As we obtain a warning that the uncertainty of the path structure sampled is very high, we stop after 100 steps. The ASE algorithm with LBFGS optimizer does not find good intermediate structures and does not converge at all. Our test showed that the FIRE optimizer works better in this context, but still takes over 500 steps to converge, and since second order methods are faster, we consider the LBFGS routine throughout this notebook.
We thus want to look at a different code, which manages to compute a NEB for this simple system more efficiently.
eOn and Metatomic¶
eOn has two improvements to accurately locate the saddle point.
Energy weighting for improving tangent resolution near the climbing image
The Off-path climbing image (6) which involves iteratively switching to the dimer method for faster convergence by the climbing image.
To use eOn, we setup a function that writes the desired eOn input for us and
runs the eonclient binary. Since we are in a notebook environment, we will
use several abstractions over raw subprocess calls. In practice, writing
and using eOn involves a configuration file, which we define as a dictionary
to be used with a helper to generate the final output.
# Define configuration as a dictionary for clarity
neb_settings = {
"Main": {"job": "nudged_elastic_band", "random_seed": 706253457},
"Potential": {"potential": "Metatomic"},
"Metatomic": {"model_path": fname.absolute()},
"Nudged Elastic Band": {
"images": N_INTERMEDIATE_IMGS,
# initialization
"initializer": "file",
"initial_path_in": "idppPath.dat",
"minimize_endpoints": "false",
# CI-NEB settings
"climbing_image_method": "true",
"climbing_image_converged_only": "true",
"ci_after": 0.5,
"ci_after_rel": 0.8,
# energy weighing
"energy_weighted": "true",
"ew_ksp_min": 0.972,
"ew_ksp_max": 9.72,
# OCI-NEB settings
"ci_mmf": "true",
"ci_mmf_after": 0.1,
"ci_mmf_after_rel": 0.5,
"ci_mmf_penalty_strength": 1.5,
"ci_mmf_penalty_base": 0.4,
"ci_mmf_angle": 0.9,
"ci_mmf_nsteps": 1000,
},
"Optimizer": {
"max_iterations": 1000,
"opt_method": "lbfgs",
"max_move": 0.1,
"converged_force": 0.01,
},
"Debug": {"write_movies": "true"},
}
Which now let’s us write out the final triplet of reactant, product, and configuration of the eOn-NEB.
write_eon_config(Path("."), neb_settings)
aseio.write("reactant.con", reactant)
aseio.write("product.con", product)
Wrote eOn config to 'config.ini'
Run the main C++ client¶
This runs ‘eonclient’ and streams output live. If it fails, the notebook execution stops here.
run_command_or_exit(["eonclient"], capture=True, timeout=300)
EON Client
VERSION: 2.10.2
BUILD DATE: Sun Feb 22 04:00:02 AM GMT 2026
OS: linux
Arch: x86_64
Hostname: runnervmwffz4
PID: 2651
DIR: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb
Loading parameter file config.ini
* [Main] job: nudged_elastic_band
* [Main] random_seed: 706253457
* [Potential] potential: Metatomic
* [Debug] write_movies: true
* [Optimizer] opt_method: lbfgs
* [Optimizer] converged_force: 0.01
* [Optimizer] max_iterations: 1000
* [Optimizer] max_move: 0.1
* [Metatomic] model_path: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/models/pet-mad-s-v1.1.0.pt
* [Nudged Elastic Band] images: 10
* [Nudged Elastic Band] energy_weighted: true
* [Nudged Elastic Band] ew_ksp_min: 0.972
* [Nudged Elastic Band] ew_ksp_max: 9.72
* [Nudged Elastic Band] climbing_image_method: true
* [Nudged Elastic Band] climbing_image_converged_only: true
* [Nudged Elastic Band] ci_after: 0.5
* [Nudged Elastic Band] ci_after_rel: 0.8
* [Nudged Elastic Band] ci_mmf: true
* [Nudged Elastic Band] ci_mmf_after: 0.1
* [Nudged Elastic Band] ci_mmf_after_rel: 0.5
* [Nudged Elastic Band] ci_mmf_nsteps: 1000
* [Nudged Elastic Band] ci_mmf_angle: 0.9
* [Nudged Elastic Band] ci_mmf_penalty_strength: 1.5
* [Nudged Elastic Band] ci_mmf_penalty_base: 0.4
* [Nudged Elastic Band] initializer: file
* [Nudged Elastic Band] initial_path_in: idppPath.dat
* [Nudged Elastic Band] minimize_endpoints: false
minimize_endpoints == false: not minimizing endpoints.
Nudged elastic band calculation started.
===============================================================
NEB Optimization Configuration
===============================================================
Baseline Force : 5.7337
Climbing Image (CI) : ENABLED
- Relative Trigger : 4.5869 (Factor: 0.80)
- Absolute Trigger : 0.5000
- Converged Only : true
Hybrid MMF (RONEB) : ENABLED
- Initial Threshold : 2.8668 (Factor: 0.50)
- Absolute Floor : 0.1000
- Penalty Scheme : 0.40 (Base: 0.40, Str: 1.50)
- Angle Tolerance : 0.9000
---------------------------------------------------------------
iteration step size ||Force|| max image max energy
---------------------------------------------------------------
1 0.0000e+00 5.7337e+00 7 1.128
2 3.2053e-02 3.8769e+00 7 0.9612
3 3.0213e-02 2.6346e+00 7 0.8915
4 8.7707e-02 2.6746e+00 7 0.7651
5 4.3540e-02 3.5079e+00 8 0.7264
6 2.2422e-02 2.0698e+00 8 0.6829
7 1.1595e-02 1.4502e+00 8 0.6673
8 4.3391e-02 2.5206e+00 8 0.6494
9 4.0043e-02 1.7329e+00 8 0.5828
10 3.3714e-02 1.2527e+00 8 0.5409
11 1.3163e-02 8.6038e-01 8 0.5278
Triggering MMF. Force: 0.8604, Threshold: 2.8668 (0.50x baseline)
Saddle point search started from reactant with energy -53.52205276489258 eV.
[Dimer] Step Step Size Delta E ||Force|| Curvature Torque Angle Rots
[IDimerRot] ----- --------- ---------- ---------- -2.4748 4.398 4.83 1 1.000
[Dimer] 1 0.0146922 -0.0061 1.03149e+00 -2.4748 4.398 4.831 1
[IDimerRot] ----- --------- ---------- ---------- -3.1533 3.106 5.82 1 0.996
[IDimerRot] ----- --------- ---------- ---------- -3.4280 2.632 8.41 2 0.986
[IDimerRot] ----- --------- ---------- ---------- -3.4819 1.970 2.52 3 0.960
[Dimer] 2 0.0095207 -0.0124 8.34226e-01 -3.4819 1.970 2.516 3
[IDimerRot] ----- --------- ---------- ---------- -3.2994 1.805 2.87 1 0.952
[Dimer] 3 0.0728679 -0.0429 7.44260e-01 -3.2994 1.805 2.871 1
[IDimerRot] ----- --------- ---------- ---------- -2.0746 2.747 1.69 1 0.942
[Dimer] 4 0.0750879 -0.0608 6.48813e-01 -2.0746 2.747 1.689 1
[IDimerRot] ----- --------- ---------- ---------- -0.8945 2.572 2.11 1 0.943
[Dimer] 5 0.0591419 -0.0616 1.14013e+00 -0.8945 2.572 2.109 1
[IDimerRot] ----- --------- ---------- ---------- -0.6226 2.395 4.37 1 0.946
[Dimer] 6 0.0137169 -0.0684 3.62446e-01 -0.6226 2.395 4.369 1
[IDimerRot] ----- --------- ---------- ---------- -0.5521 3.204 4.34 1 0.952
[Dimer] 7 0.0085159 -0.0693 2.20628e-01 -0.5521 3.204 4.343 1
[IDimerRot] ----- --------- ---------- ---------- -0.8078 2.003 4.29 1 0.952
[Dimer] 8 0.0100765 -0.0704 2.27475e-01 -0.8078 2.003 4.289 1
[IDimerRot] ----- --------- ---------- ---------- -0.9852 1.230 1.27 1 0.954
[Dimer] 9 0.0280292 -0.0720 3.03556e-01 -0.9852 1.230 1.268 1
[IDimerRot] ----- --------- ---------- ---------- -1.3565 1.640 1.12 1 0.956
[Dimer] 10 0.0266030 -0.0728 1.78914e-01 -1.3565 1.640 1.123 1
[IDimerRot] ----- --------- ---------- ---------- -1.6963 2.221 1.46 1 0.957
[Dimer] 11 0.0171125 -0.0729 1.12288e-01 -1.6963 2.221 1.462 1
[IDimerRot] ----- --------- ---------- ---------- -1.9653 1.712 3.91 1 0.957
[Dimer] 12 0.0035260 -0.0730 3.53023e-02 -1.9653 1.712 3.913 1
[IDimerRot] ----- --------- ---------- ---------- -1.8913 1.002 1.20 1 0.958
[Dimer] 13 0.0012492 -0.0731 2.66803e-02 -1.8913 1.002 1.203 1
[IDimerRot] ----- --------- ---------- ---------- -1.8976 0.947 1.98 1 0.958
[Dimer] 14 0.0018259 -0.0731 2.38627e-02 -1.8976 0.947 1.981 1
[IDimerRot] ----- --------- ---------- ---------- -1.9244 0.730 0.68 1 0.958
[Dimer] 15 0.0036333 -0.0730 3.69023e-02 -1.9244 0.730 0.681 1
[IDimerRot] ----- --------- ---------- ---------- -2.0448 0.485 0.88 1 0.958
[Dimer] 16 0.0025971 -0.0730 2.91452e-02 -2.0448 0.485 0.883 1
[IDimerRot] ----- --------- ---------- ---------- -2.1021 0.898 ------ ---- 0.959
[Dimer] 17 0.0007511 -0.0730 1.18950e-02 -2.1021 0.449 4.761 0
[IDimerRot] ----- --------- ---------- ---------- -2.1157 0.434 0.26 1 0.959
[Dimer] 18 0.0005182 -0.0730 8.32518e-03 -2.1157 0.434 0.257 1
NEB converged after MMF. Force: 0.0083
0 0.000000 0.000000 -0.001429
1 0.266349 0.026955 -0.167200
2 0.528690 0.051476 -0.069607
3 0.794314 0.073551 -0.085873
4 1.067267 0.106941 -0.138350
5 1.368503 0.167992 -0.231369
6 1.689298 0.282486 -0.457521
7 1.931783 0.447067 -0.132935
8 2.181116 0.454792 0.000911
9 2.575458 0.392960 1.029227
10 2.793776 -0.218628 2.425631
11 3.201166 -0.756573 0.000388
Found 3 extrema
Energy reference: -54.04987335205078
extrema #1 at image position 7.625245420431922 with energy 0.45529426595621914 and curvature -0.02024511157255182
extrema #2 at image position 8.520815251174174 with energy 0.4746572982503139 and curvature -0.44079793423305635
extrema #3 at image position 8.000813575746095 with energy 0.4547918766696313 and curvature 0.44079793423305635
Final state:
Nudged elastic band, successful.
Generated MMF peak 00 at position 7.625 (Energy: 0.455 eV)
Generated MMF peak 01 at position 8.521 (Energy: 0.475 eV)
Timing Information:
Real time: 3.958 seconds
User time: 7.153 seconds
System time: 0.233 seconds
CompletedProcess(args=['eonclient'], returncode=0, stdout='EON Client\nVERSION: 2.10.2\nBUILD DATE: Sun Feb 22 04:00:02 AM GMT 2026\n\nOS: linux\nArch: x86_64\nHostname: runnervmwffz4\nPID: 2651\nDIR: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb\nLoading parameter file config.ini\n* [Main] job: nudged_elastic_band\n* [Main] random_seed: 706253457\n* [Potential] potential: Metatomic\n* [Debug] write_movies: true\n* [Optimizer] opt_method: lbfgs\n* [Optimizer] converged_force: 0.01\n* [Optimizer] max_iterations: 1000\n* [Optimizer] max_move: 0.1\n* [Metatomic] model_path: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/models/pet-mad-s-v1.1.0.pt\n* [Nudged Elastic Band] images: 10\n* [Nudged Elastic Band] energy_weighted: true\n* [Nudged Elastic Band] ew_ksp_min: 0.972\n* [Nudged Elastic Band] ew_ksp_max: 9.72\n* [Nudged Elastic Band] climbing_image_method: true\n* [Nudged Elastic Band] climbing_image_converged_only: true\n* [Nudged Elastic Band] ci_after: 0.5\n* [Nudged Elastic Band] ci_after_rel: 0.8\n* [Nudged Elastic Band] ci_mmf: true\n* [Nudged Elastic Band] ci_mmf_after: 0.1\n* [Nudged Elastic Band] ci_mmf_after_rel: 0.5\n* [Nudged Elastic Band] ci_mmf_nsteps: 1000\n* [Nudged Elastic Band] ci_mmf_angle: 0.9\n* [Nudged Elastic Band] ci_mmf_penalty_strength: 1.5\n* [Nudged Elastic Band] ci_mmf_penalty_base: 0.4\n* [Nudged Elastic Band] initializer: file\n* [Nudged Elastic Band] initial_path_in: idppPath.dat\n* [Nudged Elastic Band] minimize_endpoints: false\nminimize_endpoints == false: not minimizing endpoints.\nNudged elastic band calculation started.\n===============================================================\n NEB Optimization Configuration\n===============================================================\n Baseline Force : 5.7337\n Climbing Image (CI) : ENABLED\n - Relative Trigger : 4.5869 (Factor: 0.80)\n - Absolute Trigger : 0.5000\n - Converged Only : true\n Hybrid MMF (RONEB) : ENABLED\n - Initial Threshold : 2.8668 (Factor: 0.50)\n - Absolute Floor : 0.1000\n - Penalty Scheme : 0.40 (Base: 0.40, Str: 1.50)\n - Angle Tolerance : 0.9000\n---------------------------------------------------------------\n iteration step size ||Force|| max image max energy\n---------------------------------------------------------------\n\n 1 0.0000e+00 5.7337e+00 7 1.128\n 2 3.2053e-02 3.8769e+00 7 0.9612\n 3 3.0213e-02 2.6346e+00 7 0.8915\n 4 8.7707e-02 2.6746e+00 7 0.7651\n 5 4.3540e-02 3.5079e+00 8 0.7264\n 6 2.2422e-02 2.0698e+00 8 0.6829\n 7 1.1595e-02 1.4502e+00 8 0.6673\n 8 4.3391e-02 2.5206e+00 8 0.6494\n 9 4.0043e-02 1.7329e+00 8 0.5828\n 10 3.3714e-02 1.2527e+00 8 0.5409\n 11 1.3163e-02 8.6038e-01 8 0.5278\nTriggering MMF. Force: 0.8604, Threshold: 2.8668 (0.50x baseline)\nSaddle point search started from reactant with energy -53.52205276489258 eV.\n[Dimer] Step Step Size Delta E ||Force|| Curvature Torque Angle Rots\n\n[IDimerRot] ----- --------- ---------- ---------- -2.4748 4.398 4.83 1 1.000\n[Dimer] 1 0.0146922 -0.0061 1.03149e+00 -2.4748 4.398 4.831 1\n\n[IDimerRot] ----- --------- ---------- ---------- -3.1533 3.106 5.82 1 0.996\n[IDimerRot] ----- --------- ---------- ---------- -3.4280 2.632 8.41 2 0.986\n[IDimerRot] ----- --------- ---------- ---------- -3.4819 1.970 2.52 3 0.960\n[Dimer] 2 0.0095207 -0.0124 8.34226e-01 -3.4819 1.970 2.516 3\n\n[IDimerRot] ----- --------- ---------- ---------- -3.2994 1.805 2.87 1 0.952\n[Dimer] 3 0.0728679 -0.0429 7.44260e-01 -3.2994 1.805 2.871 1\n\n[IDimerRot] ----- --------- ---------- ---------- -2.0746 2.747 1.69 1 0.942\n[Dimer] 4 0.0750879 -0.0608 6.48813e-01 -2.0746 2.747 1.689 1\n\n[IDimerRot] ----- --------- ---------- ---------- -0.8945 2.572 2.11 1 0.943\n[Dimer] 5 0.0591419 -0.0616 1.14013e+00 -0.8945 2.572 2.109 1\n\n[IDimerRot] ----- --------- ---------- ---------- -0.6226 2.395 4.37 1 0.946\n[Dimer] 6 0.0137169 -0.0684 3.62446e-01 -0.6226 2.395 4.369 1\n\n[IDimerRot] ----- --------- ---------- ---------- -0.5521 3.204 4.34 1 0.952\n[Dimer] 7 0.0085159 -0.0693 2.20628e-01 -0.5521 3.204 4.343 1\n\n[IDimerRot] ----- --------- ---------- ---------- -0.8078 2.003 4.29 1 0.952\n[Dimer] 8 0.0100765 -0.0704 2.27475e-01 -0.8078 2.003 4.289 1\n\n[IDimerRot] ----- --------- ---------- ---------- -0.9852 1.230 1.27 1 0.954\n[Dimer] 9 0.0280292 -0.0720 3.03556e-01 -0.9852 1.230 1.268 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.3565 1.640 1.12 1 0.956\n[Dimer] 10 0.0266030 -0.0728 1.78914e-01 -1.3565 1.640 1.123 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.6963 2.221 1.46 1 0.957\n[Dimer] 11 0.0171125 -0.0729 1.12288e-01 -1.6963 2.221 1.462 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.9653 1.712 3.91 1 0.957\n[Dimer] 12 0.0035260 -0.0730 3.53023e-02 -1.9653 1.712 3.913 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.8913 1.002 1.20 1 0.958\n[Dimer] 13 0.0012492 -0.0731 2.66803e-02 -1.8913 1.002 1.203 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.8976 0.947 1.98 1 0.958\n[Dimer] 14 0.0018259 -0.0731 2.38627e-02 -1.8976 0.947 1.981 1\n\n[IDimerRot] ----- --------- ---------- ---------- -1.9244 0.730 0.68 1 0.958\n[Dimer] 15 0.0036333 -0.0730 3.69023e-02 -1.9244 0.730 0.681 1\n\n[IDimerRot] ----- --------- ---------- ---------- -2.0448 0.485 0.88 1 0.958\n[Dimer] 16 0.0025971 -0.0730 2.91452e-02 -2.0448 0.485 0.883 1\n\n[IDimerRot] ----- --------- ---------- ---------- -2.1021 0.898 ------ ---- 0.959\n[Dimer] 17 0.0007511 -0.0730 1.18950e-02 -2.1021 0.449 4.761 0\n\n[IDimerRot] ----- --------- ---------- ---------- -2.1157 0.434 0.26 1 0.959\n[Dimer] 18 0.0005182 -0.0730 8.32518e-03 -2.1157 0.434 0.257 1\n\nNEB converged after MMF. Force: 0.0083\n 0 0.000000 0.000000 -0.001429\n 1 0.266349 0.026955 -0.167200\n 2 0.528690 0.051476 -0.069607\n 3 0.794314 0.073551 -0.085873\n 4 1.067267 0.106941 -0.138350\n 5 1.368503 0.167992 -0.231369\n 6 1.689298 0.282486 -0.457521\n 7 1.931783 0.447067 -0.132935\n 8 2.181116 0.454792 0.000911\n 9 2.575458 0.392960 1.029227\n 10 2.793776 -0.218628 2.425631\n 11 3.201166 -0.756573 0.000388\nFound 3 extrema\nEnergy reference: -54.04987335205078\nextrema #1 at image position 7.625245420431922 with energy 0.45529426595621914 and curvature -0.02024511157255182\nextrema #2 at image position 8.520815251174174 with energy 0.4746572982503139 and curvature -0.44079793423305635\nextrema #3 at image position 8.000813575746095 with energy 0.4547918766696313 and curvature 0.44079793423305635\nFinal state: \nNudged elastic band, successful.\nGenerated MMF peak 00 at position 7.625 (Energy: 0.455 eV)\nGenerated MMF peak 01 at position 8.521 (Energy: 0.475 eV)\nTiming Information:\n Real time: 3.958 seconds\n User time: 7.153 seconds\n System time: 0.233 seconds\n')
Visual interpretation¶
rgpycrumbs is a visualization toolkit designed to bridge the gap between raw computational output and physical intuition, mapping high-dimensional NEB trajectories onto interpretable 1D energy profiles and 2D RMSD landscapes. As it is normally used from the command-line, here we define a helper.
def run_neb_plot(
mode: str,
con_file: str = "neb.con",
output_file: str = "plot.png",
title: str = "",
rotation: str = "90x,0y,0z",
) -> list[str]:
"""
Constructs the CLI command for rgpycrumbs plotting to avoid clutter in notebooks.
mode: 'profile' (1D) or 'landscape' (2D)
"""
base_cmd = [
sys.executable,
"-m",
"rgpycrumbs.cli",
"eon",
"plt_neb",
"--con-file",
con_file,
"--output-file",
output_file,
"--ase-rotation",
rotation,
"--facecolor",
"white",
"--figsize",
"5.37",
"5.37",
"--plot-structures",
"crit_points",
]
if title:
base_cmd.extend(["--title", title])
if mode == "profile":
base_cmd.extend(
[
"--plot-type",
"profile",
"--zoom-ratio",
"0.15",
]
)
elif mode == "landscape":
base_cmd.extend(
[
"--plot-type",
"landscape",
"--rc-mode",
"path",
"--fontsize-base",
"16",
"--landscape-mode",
"surface",
"--landscape-path",
"all",
"--show-pts",
"--zoom-ratio",
"0.35",
"--surface-type",
"grad_imq",
]
)
else:
raise ValueError(f"Unknown plot mode: {mode}")
# Run the generated command
run_command_or_exit(base_cmd, capture=False, timeout=60)
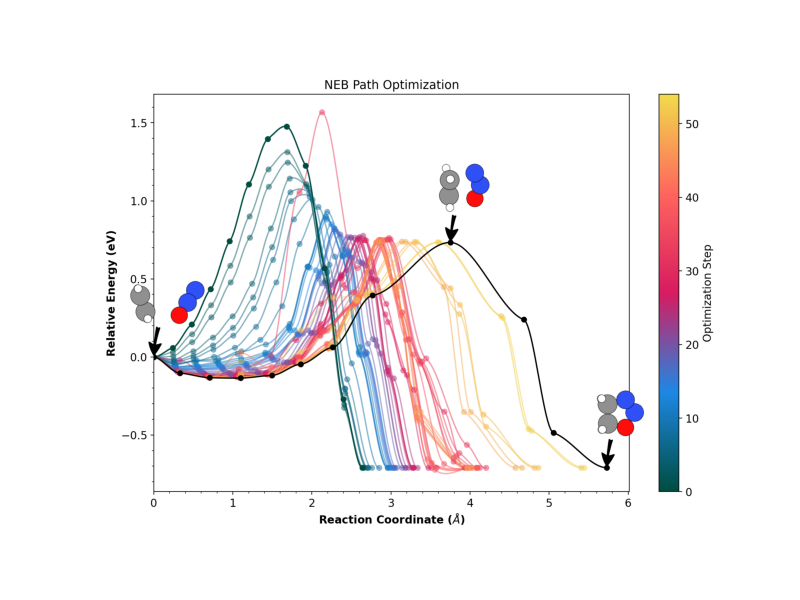
We check both the standard 1D profile against the path reaction coordinate, or the distance between intermediate images:
# Clean env to prevent backend conflicts in notebooks
os.environ.pop("MPLBACKEND", None)
# Run the 1D plotting command using the helper
run_neb_plot("profile", title="NEB Path Optimization", output_file="1D_oxad.png")
# Display the result
img = mpimg.imread("1D_oxad.png")
plt.figure(figsize=(5.37, 5.37))
plt.imshow(img)
plt.axis("off")
plt.show()
--> Dispatching to: uv run /home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/eon-pet-neb/lib/python3.13/site-packages/rgpycrumbs/eon/plt_neb.py --con-file neb.con --output-file 1D_oxad.png --ase-rotation 90x,0y,0z --facecolor white --figsize 5.37 5.37 --plot-structures crit_points --title NEB Path Optimization --plot-type profile --zoom-ratio 0.15
Downloading polars-runtime-32 (43.6MiB)
Downloading pillow (6.7MiB)
Downloading numpy (15.8MiB)
Downloading fonttools (4.7MiB)
Downloading pygments (1.2MiB)
Downloading jax (2.8MiB)
Downloading scipy (33.6MiB)
Downloading matplotlib (8.3MiB)
Downloading ml-dtypes (4.8MiB)
Downloading jaxlib (76.6MiB)
Downloading kiwisolver (1.4MiB)
Downloading ase (2.8MiB)
Downloaded kiwisolver
Downloaded pygments
Downloaded ml-dtypes
Downloaded fonttools
Downloaded pillow
Downloaded jax
Downloaded matplotlib
Downloaded polars-runtime-32
Downloaded numpy
Downloaded ase
Downloaded jaxlib
Downloaded scipy
Installed 33 packages in 68ms
[02/23/26 14:53:21] INFO INFO - Setting global rcParams for ruhi theme
WARNING WARNING - Font 'Atkinson Hyperlegible' not found.
Falling back to 'sans-serif'.
INFO INFO - Reading structures from neb.con
INFO INFO - Loaded 12 structures.
INFO INFO - Loading explicit saddle point from sp.con
INFO INFO - Searching for files with pattern:
'neb_*.dat'
INFO INFO - Found 12 file(s).
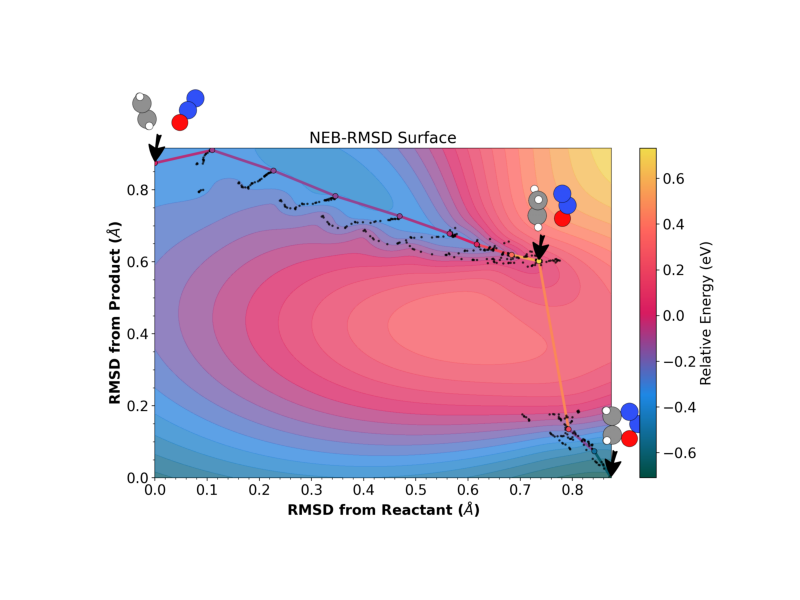
Also, the PES 2D landscape profile as a function of the RMSD [3] which shows the relative distance between the endpoints as the optimization takes place:
# Run the 2D plotting command using the helper
run_neb_plot("landscape", title="NEB-RMSD Surface", output_file="2D_oxad.png")
# Display the result
img = mpimg.imread("2D_oxad.png")
plt.figure(figsize=(5.37, 5.37))
plt.imshow(img)
plt.axis("off")
plt.show()
--> Dispatching to: uv run /home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/eon-pet-neb/lib/python3.13/site-packages/rgpycrumbs/eon/plt_neb.py --con-file neb.con --output-file 2D_oxad.png --ase-rotation 90x,0y,0z --facecolor white --figsize 5.37 5.37 --plot-structures crit_points --title NEB-RMSD Surface --plot-type landscape --rc-mode path --fontsize-base 16 --landscape-mode surface --landscape-path all --show-pts --zoom-ratio 0.35 --surface-type grad_imq
[02/23/26 14:53:24] INFO INFO - Setting global rcParams for ruhi theme
WARNING WARNING - Font 'Atkinson Hyperlegible' not found.
Falling back to 'sans-serif'.
INFO INFO - Reading structures from neb.con
INFO INFO - Loaded 12 structures.
INFO INFO - Loading explicit saddle point from sp.con
INFO INFO - Searching for files with pattern:
'neb_*.dat'
INFO INFO - Found 12 file(s).
INFO INFO - Searching for files with pattern:
'neb_path_*.con'
INFO INFO - Found 12 file(s).
INFO INFO - Computing Landscape data...
INFO INFO - Saving Landscape cache to
.neb_landscape.parquet...
INFO INFO - Calculated heuristic RBF smoothing: 0.1008
INFO INFO - Generating 2D surface using grad_imq...
INFO INFO - Optimizing hyperparameters on subset of 12
points...
INFO:2026-02-23 14:53:24,508:jax._src.xla_bridge:834: Unable to initialize backend 'tpu': INTERNAL: Failed to open libtpu.so: libtpu.so: cannot open shared object file: No such file or directory
INFO INFO - Unable to initialize backend 'tpu':
INTERNAL: Failed to open libtpu.so: libtpu.so:
cannot open shared object file: No such file or
directory
[02/23/26 14:53:30] INFO INFO - Learned Params :: LS/Eps: 0.6678, Noise:
0.0173
INFO INFO - Fitting final surface on 144 points...
[02/23/26 14:53:31] INFO INFO - Plotting explicit SP at R=0.682, P=0.609
WARNING WARNING - Looks like you are using a tranform that
doesn't support FancyArrowPatch, using ax.annotate
instead. The arrows might strike through texts.
Increasing shrinkA in arrowprops might help.
/home/runner/work/atomistic-cookbook/atomistic-cookbook/.nox/eon-pet-neb/lib/python3.13/site-packages/rgpycrumbs/eon/plt_neb.py:891: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout(pad=0.5)
Where each black dot represents a configuration at which the energy and forces are calculated during the NEB optimization run, and the RMSD is the “permutation corrected” distance from the reactant and product. The interpolated relative energy surface is generated by a Gaussian approximation with a radial basis function kernel interpolation over the energy and forces of all the black dots. See [3, Chapter 4] for more details of this visualization.
Relaxing the endpoints with eOn¶
In this final part we come back to an essential point of performing NEB calculations, and that is the relaxation of the initial states. In the tutorials above we used directly relaxed structures, and here we are demonstrating how these can be relaxed. We first load structures which are not relaxed.
reactant = aseio.read("data/reactant.con")
product = aseio.read("data/product.con")
# For compatibility with eOn, we also need to provide
# a unit cell
def center_cell(atoms):
atoms.set_cell([20, 20, 20])
atoms.pbc = True
atoms.center()
return atoms
reactant = center_cell(reactant)
product = center_cell(product)
The resulting reactant has a larger box:
fig, (ax1, ax2) = plt.subplots(1, 2)
plot_atoms(reactant, ax1, rotation=("-90x,0y,0z"))
plot_atoms(product, ax2, rotation=("-90x,0y,0z"))
ax1.text(0.3, -1, "reactant")
ax2.text(0.3, -1, "product")
ax1.set_axis_off()
ax2.set_axis_off()
# Reactant setup
dir_reactant = Path("min_reactant")
dir_reactant.mkdir(exist_ok=True)
aseio.write(dir_reactant / "pos.con", reactant)
# Product setup
dir_product = Path("min_product")
dir_product.mkdir(exist_ok=True)
aseio.write(dir_product / "pos.con", product)
# Shared minimization settings
min_settings = {
"Main": {"job": "minimization", "random_seed": 706253457},
"Potential": {"potential": "Metatomic"},
"Metatomic": {"model_path": fname.absolute()},
"Optimizer": {
"max_iterations": 2000,
"opt_method": "lbfgs",
"max_move": 0.1,
"converged_force": 0.01,
},
}
write_eon_config(dir_reactant, min_settings)
write_eon_config(dir_product, min_settings)
Wrote eOn config to 'min_reactant/config.ini'
Wrote eOn config to 'min_product/config.ini'
Run the minimization¶
The ‘eonclient’ will use the correct configuration within the folder.
@contextlib.contextmanager
def work_in_dir(path: Path):
"""
Context manager to safely change directory and return to previous
one afterwards. Crucial for notebooks to avoid path drift.
"""
prev_cwd = Path.cwd()
os.chdir(path)
try:
yield
finally:
os.chdir(prev_cwd)
with work_in_dir(dir_reactant):
run_command_or_exit(["eonclient"], capture=True, timeout=300)
with work_in_dir(dir_product):
run_command_or_exit(["eonclient"], capture=True, timeout=300)
EON Client
VERSION: 2.10.2
BUILD DATE: Sun Feb 22 04:00:02 AM GMT 2026
OS: linux
Arch: x86_64
Hostname: runnervmwffz4
PID: 2737
DIR: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/min_reactant
Loading parameter file config.ini
* [Main] job: minimization
* [Main] random_seed: 706253457
* [Potential] potential: Metatomic
* [Optimizer] opt_method: lbfgs
* [Optimizer] converged_force: 0.01
* [Optimizer] max_iterations: 2000
* [Optimizer] max_move: 0.1
* [Metatomic] model_path: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/models/pet-mad-s-v1.1.0.pt
Beginning minimization of pos.con
[Matter] Iter Step size ||Force|| Energy
[Matter] 0 0.00000e+00 2.59520e+00 -53.87877
[Matter] 1 2.66992e-02 1.68092e+00 -53.92310
[Matter] 2 1.08483e-02 4.96385e-01 -53.93404
[Matter] 3 1.38089e-03 3.48320e-01 -53.93533
[Matter] 4 6.07345e-03 1.98165e-01 -53.93784
[Matter] 5 2.36828e-03 1.59983e-01 -53.93839
[Matter] 6 3.42056e-03 1.48596e-01 -53.93892
[Matter] 7 8.11227e-03 2.15941e-01 -53.93988
[Matter] 8 2.03965e-02 2.47212e-01 -53.94168
[Matter] 9 4.77717e-02 3.64414e-01 -53.94519
[Matter] 10 1.47141e-02 3.06734e+00 -53.92278
[Matter] 11 1.27314e-02 2.21770e-01 -53.94609
[Matter] 12 7.08847e-04 2.05288e-01 -53.94638
[Matter] 13 1.48015e-02 3.05469e-01 -53.94891
[Matter] 14 9.82532e-03 6.73808e-01 -53.94783
[Matter] 15 5.09977e-03 1.50975e-01 -53.94987
[Matter] 16 1.32262e-03 1.12460e-01 -53.95013
[Matter] 17 8.90625e-03 1.63704e-01 -53.95098
[Matter] 18 9.25617e-03 2.02164e-01 -53.95173
[Matter] 19 5.29550e-02 3.43750e-01 -53.95536
[Matter] 20 1.35627e-02 1.61682e+00 -53.94640
[Matter] 21 9.79583e-03 2.62714e-01 -53.95738
[Matter] 22 1.13147e-03 1.65790e-01 -53.95789
[Matter] 23 4.29818e-03 1.66260e-01 -53.95872
[Matter] 24 4.52047e-03 1.87267e-01 -53.95936
[Matter] 25 1.26368e-02 2.55349e-01 -53.96045
[Matter] 26 5.55203e-03 1.41866e-01 -53.96107
[Matter] 27 4.08616e-03 1.59605e-01 -53.96147
[Matter] 28 1.33865e-02 2.18876e-01 -53.96261
[Matter] 29 3.77650e-02 3.29042e-01 -53.96535
[Matter] 30 8.59714e-02 7.44650e-01 -53.96917
[Matter] 31 5.04710e-02 6.64991e-01 -53.97663
[Matter] 32 3.69682e-02 4.10949e+00 -53.90312
[Matter] 33 2.87599e-02 3.14422e-01 -53.98344
[Matter] 34 2.88141e-03 2.67154e-01 -53.98453
[Matter] 35 1.37297e-02 2.19371e-01 -53.98670
[Matter] 36 4.81951e-03 4.20783e-01 -53.98696
[Matter] 37 1.91695e-03 1.98191e-01 -53.98748
[Matter] 38 2.10754e-03 1.17685e-01 -53.98796
[Matter] 39 8.15326e-03 2.31078e-01 -53.98887
[Matter] 40 1.27299e-02 2.84339e-01 -53.98985
[Matter] 41 3.07522e-02 4.83939e-01 -53.99188
[Matter] 42 3.49682e-02 3.29192e-01 -53.99386
[Matter] 43 2.49536e-02 3.07211e-01 -53.99615
[Matter] 44 4.85786e-02 5.20686e-01 -53.99976
[Matter] 45 5.88919e-03 8.85896e-01 -53.99939
[Matter] 46 3.24126e-03 2.54372e-01 -54.00129
[Matter] 47 9.70424e-04 1.91442e-01 -54.00170
[Matter] 48 5.18517e-03 1.92997e-01 -54.00282
[Matter] 49 3.45113e-03 1.64120e-01 -54.00334
[Matter] 50 1.13291e-02 2.63960e-01 -54.00426
[Matter] 51 8.49570e-03 2.45105e-01 -54.00480
[Matter] 52 1.73564e-03 1.28128e-01 -54.00521
[Matter] 53 2.45755e-03 1.14930e-01 -54.00560
[Matter] 54 5.24882e-03 1.71644e-01 -54.00615
[Matter] 55 1.62326e-02 2.76084e-01 -54.00761
[Matter] 56 3.61017e-02 3.83907e-01 -54.01062
[Matter] 57 1.37241e-02 2.41345e+00 -53.99088
[Matter] 58 9.90436e-03 2.70475e-01 -54.01271
[Matter] 59 1.10170e-03 1.82381e-01 -54.01321
[Matter] 60 5.59272e-03 1.65504e-01 -54.01412
[Matter] 61 2.55440e-03 1.80643e-01 -54.01450
[Matter] 62 3.44077e-03 1.89051e-01 -54.01492
[Matter] 63 8.38758e-03 1.67298e-01 -54.01582
[Matter] 64 1.39183e-02 2.31744e-01 -54.01682
[Matter] 65 1.17298e-02 1.78750e-01 -54.01796
[Matter] 66 1.83273e-02 1.80299e-01 -54.01976
[Matter] 67 5.39491e-02 3.59595e-01 -54.02549
[Matter] 68 1.00000e-01 1.83215e+01 -53.09299
[Matter] 69 9.47820e-02 3.75731e-01 -54.02852
[Matter] 70 1.65122e-03 3.29281e-01 -54.02928
[Matter] 71 2.44672e-02 4.45117e-01 -54.03323
[Matter] 72 8.65813e-03 7.17782e-01 -54.03351
[Matter] 73 2.98885e-03 2.91043e-01 -54.03562
[Matter] 74 3.76614e-03 1.78583e-01 -54.03678
[Matter] 75 1.69609e-02 2.60541e-01 -54.03914
[Matter] 76 1.12326e-02 2.68892e-01 -54.04039
[Matter] 77 1.97634e-02 3.72802e-01 -54.04080
[Matter] 78 4.18606e-03 1.42868e-01 -54.04169
[Matter] 79 2.28994e-03 1.41043e-01 -54.04208
[Matter] 80 8.15268e-03 1.99380e-01 -54.04312
[Matter] 81 2.14537e-02 2.13221e-01 -54.04433
[Matter] 82 1.54533e-02 2.50699e-01 -54.04527
[Matter] 83 1.43606e-02 9.90599e-02 -54.04632
[Matter] 84 5.22693e-03 8.29255e-02 -54.04667
[Matter] 85 6.77061e-03 8.62247e-02 -54.04701
[Matter] 86 1.64103e-02 1.74262e-01 -54.04739
[Matter] 87 5.21523e-03 8.45465e-02 -54.04772
[Matter] 88 2.33663e-03 7.25062e-02 -54.04788
[Matter] 89 5.62010e-03 7.64859e-02 -54.04821
[Matter] 90 1.49492e-02 8.00489e-02 -54.04877
[Matter] 91 4.39810e-02 8.26332e-01 -54.04655
[Matter] 92 2.65137e-02 7.55921e-02 -54.04941
[Matter] 93 3.01922e-03 6.11931e-02 -54.04952
[Matter] 94 8.64540e-03 5.84156e-02 -54.04972
[Matter] 95 2.67894e-03 6.71808e-02 -54.04970
[Matter] 96 1.36744e-03 1.40823e-02 -54.04976
[Matter] 97 3.81149e-04 1.28090e-02 -54.04976
[Matter] 98 1.08826e-03 1.76208e-02 -54.04977
[Matter] 99 2.16379e-03 2.17760e-02 -54.04979
[Matter] 100 6.85528e-03 4.67164e-02 -54.04983
[Matter] 101 6.47437e-03 2.67711e-02 -54.04986
[Matter] 102 2.97434e-03 1.99054e-02 -54.04988
[Matter] 103 1.13123e-03 1.91474e-02 -54.04988
[Matter] 104 4.35915e-04 1.46577e-02 -54.04989
[Matter] 105 1.30106e-03 1.99583e-02 -54.04990
[Matter] 106 3.36394e-03 2.06340e-02 -54.04992
[Matter] 107 4.31728e-03 2.90261e-02 -54.04992
[Matter] 108 8.35306e-04 1.07014e-02 -54.04993
[Matter] 109 5.74216e-04 3.30885e-03 -54.04993
Minimization converged within tolerence
Saving result to min.con
Final Energy: -54.049930572509766
Timing Information:
Real time: 4.584 seconds
User time: 8.404 seconds
System time: 0.231 seconds
EON Client
VERSION: 2.10.2
BUILD DATE: Sun Feb 22 04:00:02 AM GMT 2026
OS: linux
Arch: x86_64
Hostname: runnervmwffz4
PID: 2740
DIR: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/min_product
Loading parameter file config.ini
* [Main] job: minimization
* [Main] random_seed: 706253457
* [Potential] potential: Metatomic
* [Optimizer] opt_method: lbfgs
* [Optimizer] converged_force: 0.01
* [Optimizer] max_iterations: 2000
* [Optimizer] max_move: 0.1
* [Metatomic] model_path: /home/runner/work/atomistic-cookbook/atomistic-cookbook/examples/eon-pet-neb/models/pet-mad-s-v1.1.0.pt
Beginning minimization of pos.con
[Matter] Iter Step size ||Force|| Energy
[Matter] 0 0.00000e+00 1.81086e+00 -54.74133
[Matter] 1 3.53440e-02 1.14322e+00 -54.78707
[Matter] 2 1.14535e-02 3.97479e-01 -54.80053
[Matter] 3 9.53215e-03 2.21687e-01 -54.80446
[Matter] 4 6.45762e-03 1.01501e-01 -54.80579
[Matter] 5 1.86501e-03 5.29620e-02 -54.80604
[Matter] 6 2.50612e-03 4.05540e-02 -54.80619
[Matter] 7 1.92328e-03 2.81717e-02 -54.80624
[Matter] 8 1.55185e-03 1.87605e-02 -54.80627
[Matter] 9 4.02005e-04 1.11695e-02 -54.80627
[Matter] 10 4.61101e-04 7.53657e-03 -54.80628
Minimization converged within tolerence
Saving result to min.con
Final Energy: -54.80628204345703
Timing Information:
Real time: 1.480 seconds
User time: 2.291 seconds
System time: 0.133 seconds
Additionally, the relative ordering must be preserved, for which we use IRA [4].
reactant = aseio.read(dir_reactant / "min.con")
product = aseio.read(dir_product / "min.con")
ira = ira_mod.IRA()
# Default value
kmax_factor = 1.8
nat1 = len(reactant)
typ1 = reactant.get_atomic_numbers()
coords1 = reactant.get_positions()
nat2 = len(product)
typ2 = product.get_atomic_numbers()
coords2 = product.get_positions()
print("Running ira.match to find rotation, translation, AND permutation...")
# r = rotation, t = translation, p = permutation, hd = Hausdorff distance
r, t, p, hd = ira.match(nat1, typ1, coords1, nat2, typ2, coords2, kmax_factor)
print(f"Matching complete. Hausdorff Distance (hd) = {hd:.6f} Angstrom")
# Apply rotation (r) and translation (t) to the original product coordinates
# This aligns the product's orientation to the reactant's
coords2_aligned = (coords2 @ r.T) + t
# Apply the permutation (p)
# This re-orders the aligned product atoms to match the reactant's atom order
# p[i] = j means reactant atom 'i' matches product atom 'j'
# So, the new coordinate array's i-th element should be coords2_aligned[j]
coords2_aligned_permuted = coords2_aligned[p]
# Save the new aligned-and-permuted structure
# CRUCIAL: Use chemical symbols from the reactant,
# because we have now permuted the product coordinates to match the reactant order.
product = reactant.copy()
product.positions = coords2_aligned_permuted
Running ira.match to find rotation, translation, AND permutation...
Matching complete. Hausdorff Distance (hd) = 1.925488 Angstrom
Finally we can visualize these with ASE.
view(reactant, viewer="x3d")
view(product, viewer="x3d")
fig, (ax1, ax2) = plt.subplots(1, 2)
plot_atoms(reactant, ax1, rotation=("-90x,0y,0z"))
plot_atoms(product, ax2, rotation=("-90x,0y,0z"))
ax1.text(0.3, -1, "reactant")
ax2.text(0.3, -1, "product")
ax1.set_axis_off()
ax2.set_axis_off()
References¶
Mazitov, A.; Bigi, F.; Kellner, M.; Pegolo, P.; Tisi, D.; Fraux, G.; Pozdnyakov, S.; Loche, P.; Ceriotti, M. PET-MAD, a Universal Interatomic Potential for Advanced Materials Modeling. arXiv March 18, 2025. https://doi.org/10.48550/arXiv.2503.14118.
Bigi, F.; Abbott, J. W.; Loche, P.; Mazitov, A.; Tisi, D.; Langer, M. F.; Goscinski, A.; Pegolo, P.; Chong, S.; Goswami, R.; Chorna, S.; Kellner, M.; Ceriotti, M.; Fraux, G. Metatensor and Metatomic: Foundational Libraries for Interoperable Atomistic Machine Learning. arXiv August 21, 2025. https://doi.org/10.48550/arXiv.2508.15704.
Goswami, R. Efficient Exploration of Chemical Kinetics. arXiv October 24, 2025. https://doi.org/10.48550/arXiv.2510.21368.
Gunde, M.; Salles, N.; Hémeryck, A.; Martin-Samos, L. IRA: A Shape Matching Approach for Recognition and Comparison of Generic Atomic Patterns. J. Chem. Inf. Model. 2021, 61 (11), 5446–5457. https://doi.org/10.1021/acs.jcim.1c00567.
Smidstrup, S.; Pedersen, A.; Stokbro, K.; Jónsson, H. Improved Initial Guess for Minimum Energy Path Calculations. J. Chem. Phys. 2014, 140 (21), 214106. https://doi.org/10.1063/1.4878664.
Goswami, R; Gunde, M; Jónsson, H. Enhanced climbing image nudged elastic band method with hessian eigenmode alignment, Jan. 22, 2026, arXiv: arXiv:2601.12630. doi: 10.48550/arXiv.2601.12630.
Total running time of the script: (1 minutes 4.906 seconds)